Rapid chemical ligation of oligonucleotides by the Diels–Alder reaction†‡§
Afaf H.
El-Sagheer
*ab,
Vee Vee
Cheong
c and
Tom
Brown
*a
aSchool of Chemistry, University of Southampton, Highfield, Southampton, U.K. SO17 1BJ. E-mail: ahes@soton.ac.uk; tb2@soton.ac.uk
bChemistry Branch, Department of Science and Mathematics, Faculty of Petroleum and Mining Engineering, Suez Canal University, Suez, Egypt
cDivision of Chemistry and Biological Chemistry, School of Physical and Mathematical Sciences, Nanyang Technological University, Singapore 637371
First published on 11th November 2010
Abstract
Extremely fast and efficient Diels–Alder chemical ligation of furan and maleimide oligonucleotides has been carried out in aqueous buffer. It was possible to ligate three oligonucleotides simultaneously in a controlled manner with the aid of a complementary splint. The templated reactions proceeded within 1 min at room temperature whereas non-templated reactions were slow and incomplete. Rapid and clean methods of DNA ligation such as the one demonstrated here have potential uses in biology and nanotechnology.
Introduction
Clean and efficient methods of linking and cyclising DNA strands are of potential value in biology. Chemical ligation is particularly interesting in this context as it can be carried out on a large scale under a variety of conditions and it does not depend on the use of enzymes. The unnatural backbone linkages that are produced by chemical ligation need not be incompatible with biological processes. For example, it has been shown that a triazole linkage produced by the copper-catalysed alkyne-azide cycloaddition (CuAAC) reaction between alkyne and azide-labelled oligodeoxynucleotides (ODNs) can be read through by thermostable DNA polymerases during PCR.1 This suggests that the assembly of large DNA constructs by purely chemical methods and their conversion to natural DNA by endogenous polymerases in vivo is a distinct possibility. Another application that could be envisaged is the selective inhibition of gene expression by antisense ODN ligation using the host target nucleic acid (e.g. m-RNA, si-RNA) as a template. The use of chemical ligation in the field of nanotechnology has also been demonstrated.2 Large addressable double stranded cyclic DNA constructs (mol wt > 40 K) have been assembled and fixed using click chemistry. Purification under denaturing conditions produced building blocks for use in nano-assembly. One potential advantage of chemical ligation in bio- and nano-related applications is the diversity of chemical reactions that can be used. Synthetic chemistry also allow for orthogonality, i.e. different reactions can be carried out in parallel in a controlled manner. In order to achieve the objectives outlined above we have carried out detailed studies on one particular chemical approach, the CuAAC reaction (click-ligation).3–5 Its major limitation is the requirement for CuI-catalysis, which is difficult to achieve in vivo. The non-catalysed reaction (AAC reaction) is many orders of magnitude slower but can be accelerated under specific conditions.6 Detailed studies are being carried out to find methods of increasing its reaction rate further, but it cannot alone provide the desired orthogonality.Therefore we are currently investigating other reactions for the chemical ligation of nucleic acids. The Diels–Alder [4 + 2] cycloaddition reaction is particularly attractive for several reasons: it is well understood, it is known to proceed rapidly in water,7 it does not require catalysis, and the requisite reactants (ODNs labelled with dienes and dienophiles) are readily accessible. The Diels–Alder reaction has previously been used in the nucleic acids field for the synthesis of ODN-peptide conjugates8,9 and for ODN labelling10–12 (fluorescent and SERS)13 Treatment of 1,3-butadiene-modified complementary ODNs with difunctional dienophiles has been shown to lead to the slow formation of inter-strand crosslinks. The process is slow (7 days) because the initial Diels–Alder reactions is not templated.14 Diels–Alder chemistry has also been used to immobilise oligonucleotides on surfaces.15
Results and discussion
The objective of this study was to establish methods for the simultaneous ligation of several ODNs against a complementary DNA template (splint) in a controlled manner with the eventual aim of building large DNA constructs (Scheme 1a). As a proof of principle we initially investigated the ligation of two ODNs (Scheme 1b). Suitable amino-modified ODNs were labelled at the 5′- or 3′- ends with 3(2-furyl) propanoic acid NHS ester16 or 6-maleimidohexanoic acid NHS ester (Scheme 2) to produce chemically compatible pairs, (3′-maleimide + 5′-furan or 3′-furan + 5′-maleimide) (Table 1), which could be reacted together whilst captured on a complementary ODN template.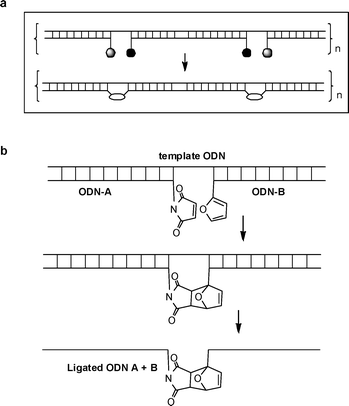 | ||
| Scheme 1 Oligonucleotide strand ligation using Diels–Alder chemistry. a. Schematic of multiple simultaneous ligations. b. Templated chemical reaction between maleimide and furan ODNs and resultant ligated single strand. ODN = oligodeoxyribonucleotide. | ||
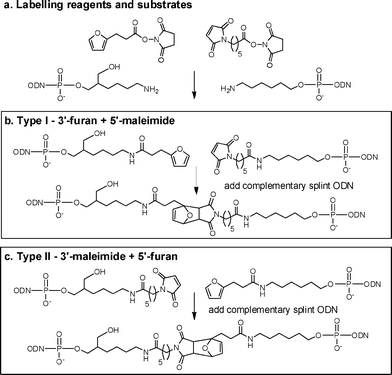 | ||
| Scheme 2 a: Furan and maleimide labelling reagents and amino-modified ODN substrates. b, c: Diels–Alder reactions to form type I and II linkages. | ||
| Entry | Sequences (5′ – 3′) |
|---|---|
| MO1 | M GCGATCAATCAGACG |
| FO2 | CTTTCCTCCACTGTTGCF |
| TO3 | TCTGATTGATCGCGCAACAGTGGAGG |
| LO4 | CTTTCCTCCACTGTTGCF+MGCGATCAATCAGACG |
| MO5 | CTTTCCTCCACTGTTGCM |
| FO6 | F GCGATCAATCAGACG |
| TO7 | ATTGATCGCGCAACAGTG |
| LO8 | CTTTCCTCCACTGTTGCM+FGCGATCAATCAGACG |
| CO9 | CTTTCCTCCACTGTTGCGCGATCAATCAGACG |
| FFO10 | F GAACTCTACGACGTAGF |
| TO11 | TTTTTTTTCGTCTGATTGATCGCCTACGTCGTAGAGTTCGCAACAGTGGAGGAAAGTTTTTTTT |
| LO12 | CTTTCCTCCACTGTTGCM+FGAACTCTACGACGTAGF +MGCGATCAATCAGACG |
One limitation of the use of the maleimide functionality is its potential instability to aqueous buffer. Detailed investigations indicated that elevated temperature, high pH (>7.5) and prolonged storage in water should be avoided (ESI‡). Taking this into account, a type I Diels–Alder linkage was created between two ODNs by the template-mediated reaction of 5′-maleimide MO1 with 3′-furan FO2 at pH 6.2 and room temperature in the presence of complementary TO3 to give ligation product LO4 (Scheme 2b).¶ Analysis of the time course of the reaction by polyacrylamide gel electrophoresis (Fig. 1) indicated that there was no significant increase in product after 1 min, indicating that extensive ligation occurred in a few seconds. Purified LO4 was characterized by mass spectrometry (MALDI-TOF [M+H]+ calc. 10352; found 10353.2).
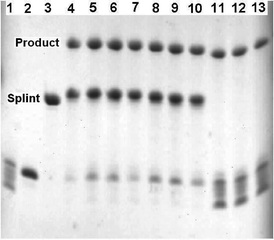 | ||
| Fig. 1 Templated and non-templated type I Diels–Alder reaction to ligate two ODNs. Lanes 1–3, participating ODNs. 1) 5′-Maleimide-labelled MO1. 2) 3′-Furan-labelled FO2. 3) Complementary template (splint) TO3. Lanes 4–7: Templated reaction to assemble ligated LO4 in 10 mM phosphate, 0.2 M NaCl, pH 6.2 buffer: 4) 1 min, 5) 15 min, 6) 30 min, 7) 1 h. Lanes 8–10: Templated reaction to assemble ligated LO4 in 10 mM phosphate, 0.2 M NaCl, pH 7.0 buffer: 8) 1 min, 9) 15 min, 10) 1 h. Lanes 11–13: Non-templated reaction to assemble ligated LO4 in 10 mM phosphate, 0.2 M NaCl, pH 6.2 buffer: 11) 1 min, 12) 1 h, 13) 6 h. Maleimide ODN MO1 in lane 1 is unstable to the TBE gel buffer (pH 8.2) as confirmed by mass spectrometry (ESI‡). All ODN concentrations are 60 μmolar. Analysis by 20% denaturing polyacrylamide gel. | ||
Next, a study was carried out to determine whether the orientation of Diels–Alder linkage would affect the reaction rate. Incubation of 3′-maleimide MO5 and 5′-furan FO6 with their complementary template TO7 gave the type II Diels–Alder ligated LO8 (Scheme 2c). The reaction was conducted as described for the type I linkage and again it was very fast, with a considerable amount of Diels–Alder product formed within the first minute (Fig. 2 lanes 3–6). Characterization of the product excised from a denaturing polyacrylamide gel by mass spectrometry confirmed it to be LO8 ([M+H]+ calc. 10352; found 10353.9).
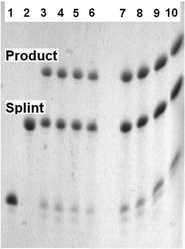 | ||
| Fig. 2 Templated type II Diels–Alder reaction to ligate two ODNs. Lane 1) 5′-furan-labelled FO6. 2) complementary template (splint) TO7. Lanes 3–6: Templated reaction to assemble ligated LO8 in 10 mM phosphate, 0.2 M NaCl, pH 6.2 buffer: 3) 1 min, 4) 15 min, 5) 30 min, 6) 1 h. Lanes 7–10: Templated reaction to assemble ligated LO8 in 10 mM phosphate, 0.2 M NaCl, pH 7.0 buffer: 7) 1 min, 8) 15 min, 9) 30 min, 10) 1 h. All ODN concentrations are 60 μmolar. Analysis by 20% denaturing polyacrylamide gel. | ||
To verify that the DNA-templated Diels–Alder reaction is viable in conditions similar to those encountered in vivo, type I and II ligations were also carried out successfully at pH 7 (Fig. 1 lanes 8–10, Fig. 2 lanes 7–10). It is presumed that the use of a complementary template brings the reacting ODNs together in a favourable orientation and thereby accelerates the reaction. To verify the importance of DNA templating, the type I and II Diels–Alder reactions were carried out in the absence of a complementary splint with extended reaction times of up to 20 h (Fig. 1 lanes 11–13 and ESI‡). The reaction was much slower than the templated version and was incomplete at the end of the full time course. UV melting studies were carried out on duplexes containing a Diels–Alder linkage in one strand. LO8 and its complementary template TO7 was found to have a Tm of 57.9 °C compared to 67.1 °C for the unmodified duplex (CO9/TO7). The type I Diels–Alder product had a slightly higher Tm (59.0 °C) than the type II analogue (ESI‡). There is likely to be scope for increasing the Tm of the Diels–Alder duplex by changing the length of the linkages between the furan/maleimide and the ODN but this has not yet been explored.
Having successfully ligated two ODNs, we investigated the simultaneous Diels–Alder ligation of three ODNs, the central one bearing furan moieties at each end. Incubation of 5′,3′-dual-furan FFO10 with 3′-maleimide MO5 and 5′-maleimide MO1 in the presence of complementary template TO11 in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio gave the desired 48-mer Diels–Alder product LO12 ([M+H]+ calc. 15946; found 15946). It was identified as the second band from the top of the gel (lanes 3–10, Fig. 3a) running just in front of the 64-mer splint TO11. The chemical structures at the ligation sites are shown in Fig. 3b.
1 ratio gave the desired 48-mer Diels–Alder product LO12 ([M+H]+ calc. 15946; found 15946). It was identified as the second band from the top of the gel (lanes 3–10, Fig. 3a) running just in front of the 64-mer splint TO11. The chemical structures at the ligation sites are shown in Fig. 3b.
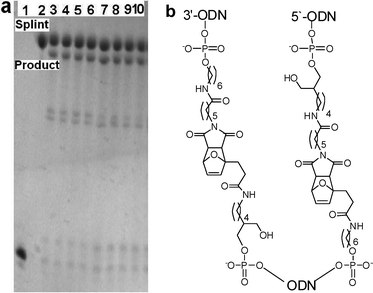 | ||
| Fig. 3 a: Type III Diels–Alder reaction to simultaneously ligate three ODNs (3′-maleimide MO5 + 5′-furan-3′-furan FFO10 + 5′-maleimide MO1). Lane 1) dual-furan-labelled FFO10. 2) Complementary template TO11. Lanes 3–6: Ligation reaction without pre-annealing of FFO10 to TO11, 3) 1 min, 4) 15 min, 5) 30 min, 6) 1 h. Lanes 7–10: Ligation reaction after heating FFO10 with TO11 at 80 °C for 5 min then cooling to RT over 2 h: 7) 1 min, 8) 15 min, 9) 30 min, 10) 1 h. All reactions carried out in 10 mM phosphate, 0.2 M NaCl, pH 6.2 buffer. All ODN concentrations are 20 μmolar. Analysis by 20% denaturing polyacrylamide gel. b: Chemical structures at ligation points. | ||
There are also two weak bands corresponding to the Diels–Alder products of only one ligation. As previously the reaction was very fast with no discernible change after the initial 1 min reaction time, and only faint traces of the maleimide and furan-labelled ODNs remained unreacted. There was a small improvement when the furan ODN was heated and cooled in the presence of the template ODN before addition of the maleimide ODNs, but both methods gave good results. These experiments establish the principle that Diels–Alder ligation can be used to join several ODNs together.
Conclusions
This study demonstrates the speed, efficiency and simplicity of the templated Diels–Alder DNA ligation reaction, which is a potentially valuable tool in nucleic acids chemistry. The success of the controlled simultaneous ligation of three oligonucleotides suggests that significantly larger constructs could be assembled by this methodology. It is also likely that the same chemistry can be applied to biologically important RNA analogues. There is also potential to vary the nature of the diene and dienophile in order to change the chemical structure at the site of ligation. This could be an important parameter in future biological applications. Work is in progress to determine the behaviour in live cells of DNA constructs that contain Diels–Alder linkages, and to pursue the use of this chemistry in nanotechnology applications involving DNA.Acknowledgements
The research leading to these results has received funding from the European Community's Seventh Framework Programme (FP7/2007–2013) under Grant No. HEALTH-F4-2008-201418, entitled READNA.Notes and references
- A. H. El-Sagheer and T. Brown, J. Am. Chem. Soc., 2009, 131, 3958–3964 CrossRef CAS.
- E. P. Lundberg, A. H. El-Sagheer, P. Kocalka, M. Wilhelmsson, T. Brown and B. Norden, Chem. Commun., 2010, 46, 3714–3716 RSC.
- R. Kumar, A. El-Sagheer, J. Tumpane, P. Lincoln, L. M. Wilhelmsson and T. Brown, J. Am. Chem. Soc., 2007, 129, 6859–6864 CrossRef CAS.
- A. H. El-Sagheer, R. Kumar, S. Findlow, J. M. Werner, A. N. Lane and T. Brown, ChemBioChem, 2008, 9, 50–52 CrossRef CAS.
- P. Kocalka, A. H. El-Sagheer and T. Brown, ChemBioChem, 2008, 9, 1280–1285 CrossRef CAS.
- A. H. El-Sagheer and T. Brown, Pure Appl. Chem., 2010, 82, 1599–1607 CrossRef CAS.
- D. C. Rideout and R. Breslow, J. Am. Chem. Soc., 1980, 102, 7816–7817 CrossRef CAS.
- V. Marchan, S. Ortega, D. Pulido, E. Pedroso and A. Grandas, Nucleic Acids Res., 2006, 34, e24 CrossRef.
- V. Steven and D. Graham, Org. Biomol. Chem., 2008, 6, 3781–3787 RSC.
- V. Borsenberger and S. Howorka, Nucleic Acids Res., 2009, 37, 1477–1485 CrossRef CAS.
- D. Graham, A. Grondin, C. McHugh, L. Fruk and W. E. Smith, Tetrahedron Lett., 2002, 43, 4785–4788 CrossRef CAS.
- L. Fruk, A. Grondin, W. E. Smith and D. Graham, Chem. Commun., 2002, 2100–2101 RSC.
- D. Graham, L. Fruk and W. E. Smith, Analyst, 2003, 128, 692–699 RSC.
- R. Tona and R. Haner, Mol. BioSyst., 2005, 1, 93–98 RSC.
- G. M. Husar, D. J. Anziano, M. Leuck and D. P. Sebesta, Nucleosides, Nucleotides Nucleic Acids, 2001, 20, 559–566 CrossRef CAS.
- L. S. Huang and R. J. Kerns, Bioorg. Med. Chem., 2006, 14, 2300–2313 CrossRef CAS.
Footnotes |
| † This paper is part of an Organic & Biomolecular Chemistry web theme issue on chemical biology. |
| ‡ Electronic supplementary information (ESI) available: Experimental details, UV melting studies, ODN mass spectral data, maleimide ODN stability studies. See DOI: 10.1039/c0ob00451k |
| § T.B and A.H. E-S are joint main authors. |
| ¶ Throughout this study all Diels–Alder reactions were stopped at the designated time point by dilution with water followed by rapid gel-filtration, then immediate freezing in liquid N2. The timescale for this process was 1 min. The frozen samples were lyophilised prior to gel-electrophoresis. |
| This journal is © The Royal Society of Chemistry 2011 |
