DOI:
10.1039/C0MD00200C
(Concise Article)
Med. Chem. Commun., 2011,
2, 132-139
New positive allosteric modulators of the metabotropic glutamate receptor 2 (mGluR2). Identification and synthesis of N-propyl-5-substituted isoquinolones
Received
3rd November 2010
, Accepted 30th November 2010
First published on 17th December 2010
Introduction
Activation of group II metabotropic COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundglutamate receptors (mGluR) mGluR2 and mGluR3 may provide anxiolytic and/or antipsychotic effects.1,2,3 The mixed mGluR2/mGluR3 agonist LY354740 (1, Fig. 1) has shown anxiolytic potential in healthy human volunteers, showing activity in fear-potentiated startle and panic induction after CO2 challenge.4,5 A related prodrug LY2140023 (2) demonstrated improvements in positive and negative symptoms compared to placebo in schizophrenic patients.6 There is evidence from mice knock-out studies that preclinical anti-psychotic effects may be mediated via the mGluR2 receptor.7
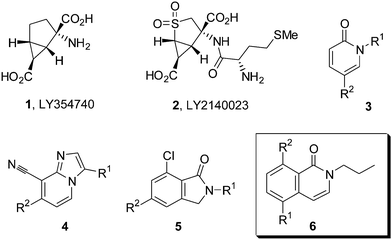 |
| | Fig. 1 Structures of mixed mGluR2/3 agonists LY354740 (1), LY2140023 (2), and recent mGluR2 PAM chemotypes (3–5). | |
There is increasing interest in identifying positive allosteric modulators (PAMs) of mGluR2 which bind at an alternative site to the orthosteric endogenous agonist.8 We have recently reviewed the currently known mGluR2 PAM chemotypes9 and new reports are appearing on a frequent basis.10,11,12 The general structures of three of those recently disclosed mGuR2PAM chemotypes 3–5 are shown in Fig. 1. In this letter we present the discovery and preliminary SAR exploration of N-propyl-5-substituted isoquinolones 6 as a new series of mGluR2 PAMs.
Results and discussion
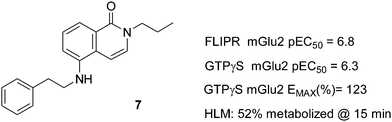
High-throughput screening of the Addex Pharmaceuticals compound collection in an mGluR2 PAMFLIPR (fluorometric imaging plate reader) assay resulted in the discovery of a singleton COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundisoquinolone7 which possessed an interesting functional potency (FLIPR pEC50 = 6.8). The potentiating activity of 7 was confirmed via a subsequent mGluR2 [35S]-GTPγS assay (Fig. 2). Two different read outs were obtained from the GTPγS assay: (a) pEC50 for potentiation of the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundglutamate signal, and (b) the maximal response obtained using the test compound plus an EC20 of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundglutamate normalized to the maximal response obtained with COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundglutamate alone (% EMAX)13 Compound 7 had a GTPγS pEC50 of 6.3 and an EMAX of 123%. 7 was tested for single point microsomal stability in human liver microsomes (HLM) and showed moderate metabolic stability (52% metabolized after 15 min incubation). We considered the singleton 7 as a potentially attractive starting point for an initial limited CNS focused SAR exploration aiming to confirm activity within the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundisoquinolone chemotype 6.
The initial set of compounds 7–20 covered variation of the R1group at position C-5 of the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundisoquinolone ring while maintaining the R2 substituent constant and equal to COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundhydrogen (R2 = H). Some examples where R2 = COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCl (COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound21–23) were also designed to study the similarity of this series with the reported COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundisoindolone derivatives 5 (Fig. 1).
Chemistry
The target compounds 7–23 were prepared following the synthetic strategies shown in Schemes 1–3.
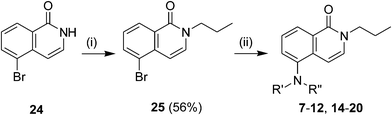 |
| | Scheme 1 Preparation of 6-aminoisoquinolones. Reagents and conditions: (i) COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundNaH, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound1-bromopropane, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundDMF, 0 °C to rt, 12 h. (ii) R'R′′NH, t-BuONa, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundBINAP, Pd2(dba)3, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundtoluene, μW, 180 °C, 1 h, 12–98%. | |
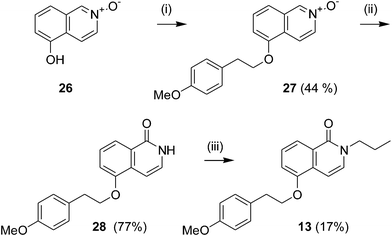 |
| | Scheme 2 Preparation of 6-alkoxyisoquinolones. Reagents and conditions: (i) COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound1-(2-chloroethyl)-4-methoxybenzene, K2CO3, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCH3CN, 180 °C μW, 15 min. (ii) a) Ac2O, 120 °C, 3 h; b) COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundNaOH 2 M, 50 °C, 1 h. (iii) COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundNaH, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound1-bromopropane, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundDMF, 0 °C to rt, 12 h. | |
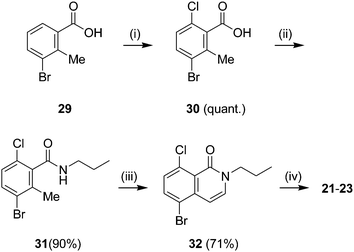 |
| | Scheme 3 Preparation of 8-chloro-substituted isoquinolones. Reagents and conditions: (i) NCS, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundPd(OAc)2, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundDMF, 110 °C, μW, 15 min. (ii) COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundN-propylamine, EDCI, HOBt, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundDCM, 50 °C, 8 h. (iii) COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundLDA, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundDMF, −78 °C to −10 °C, 12 h. (iv) RNH2, t-BuONa, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundBINAP, Pd2(dba)3, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundtoluene, 110 °C, 2 h, 40–72%. | |
The majority of final compounds were synthesized from commercially available COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound5-bromoisoquinolone24 following the synthesis steps shown in Scheme 1. Thus N-alkylation of 24 with N-bromopropane in the presence of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundsodium hydride afforded compound COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound25 in 56% yield. Microwave-assisted Buchwald–Hartwig type cross-coupling reaction of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound25 with several amines yielded the corresponding coupling products in variable yield (12–98%) depending on the nature of the coupled amine.
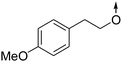
Compound 13, bearing an ether linker, was prepared as shown in Scheme 2. O-alkylation of the N-oxide derivative 26 with COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound4-methoxyphenethyl bromide afforded the N-oxide27 in moderate yield.14 Heating of compound 27 in COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundacetic anhydride gave an acyl derivative that was not isolated but converted directly to the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundisoquinolone28 by treatment with COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundNaOH.15N-alkylation of 28 with COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound1-bromopropane afforded the final compound 13 in 17% yield.
Finally, compounds COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound21–23 bearing a COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundchlorine substituent at position C-8 were prepared as shown in Scheme 3. Commercially available COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound3-bromo-2-methylbenzoic acid29 was chlorinated by treatment with COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundN-chlorosuccinimide (NCS) in the presence of 0.3 to 0.8 equivalents of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundPd(OAc)2 to yield compound COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound30,16 which was then transformed into the amideCOMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound31 under standard amide formation conditions. Ring cyclisation of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound31 by reaction with COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundLDA and subsequest quenching of the benzylic organolithium with COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundDMF afforded COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound8-chloro-5-bromo-N-propylisoquinoloneCOMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound32 in good yield.17 Buchwald–Hartwig type coupling of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound32 with the corresponding amines afforded the targeted compounds COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound21–23.
Pharmacology
The variations around the R1 and R2groups and the functional activity and metabolic stability data in human liver microsomes (HLM) of N-propylisoquinolones 7–23 are listed in Table 1.18
Table 1 GTPγS functional activity and HLM stability data of representative mGluR2 PAMs 7–23
Compounds 7–20 were the result of the exploration around R1 while keeping constant R2 = H. Firstly, the effect of different substituents on the distal phenyl ring was explored. Thus, a methoxy substituent (COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound8, 9 and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound10) resulted in a slight increase in potency, particularly pronounced when placed at the ortho position (pEC50 = 6.9, EMAX = 137%). Unfortunately, the stability of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound8 and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound10 in HLM worsened and both compounds showed a high metabolic turnover (COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound8: 91% and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound10 : 97% metabolized after 15 min in HLM). A 4-methyl substituent 11 was also beneficial for potency (pEC50 = 6.7, EMAX = 134%). This potency increase obtained with compounds 8–11 gave us confidence on the viablity of the singleton hit 7.
The role of the NH group at position C-5 of the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundisoquinolone ring was studied with the preparation of compounds COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound12 and 13. The activity found for both compounds was in the same range as the one of compound 7, indicating that a hydrogen donorgroup is not essential for activity at position C-5.
The distance between the distal phenyl group and the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundisoquinolone core was then explored with compounds COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound14 and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound15. A direct relationship between the length of the alkyl chain and the increase in potency was observed. Thus, phenylpropyl derivative COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound14 turned out to be half a log unit more potent than the phenethyl hit 7, with the shorter benzyl derivative COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound15, being only weakly active. Unfortunately, this potency increase observed with the propyl spacer was not accompanied by an improvement on the metabolic stability (COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound14: 57% metabolized), and only the shorter benzyl was found to be metabolically more stable (COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound15: 38% metabolized).
In view of the better metabolic profile obtained with the benzyl analogue, further mapping of the phenyl group in COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound15 was performed in an attempt to find molecules that would combine metabolic stability with high potency (16–20). Overall, the SAR on the aryl closely paralleled that of the phenethyl analogues. Thus the methoxy- derivatives COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound16 and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound17 showed comparable potency to their corresponding phenethyl pairs 9 and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound10. Compound COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound18, bearing the liphophilic but bulky phenoxy substituent was less active and had a much lower EMAX value. The best results were obtained with the small and lipophilic 3-trifluoromethoxy- (19: pEC50 = 6.7, EMAX = 162%) and chloro-analogues (COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound20: pEC50 = 6.70, EMAX = 163%). Remarkably, metabolic stability was also improved with 19 and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound20.
Given the structural analogy of our COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundisoquinolone series with reported mGluR2 PAM isoindolones, analogues COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound21–23 having a chloro atom at position C-8 (R2 = COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCl) were prepared. Unlike with the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundisoindolone series, the presence of the 8-chloro in the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundisoquinolone core was not beneficial for activity, and compounds COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound21–23 showed activity in the same range as their analogues 7, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound15 and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound16, however a significantly lower EMAX was measured in all cases.
Metabolite identification studies indicated that in most cases oxidative N-dealkylation at position C-5 of the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundisoquinolone ring is the main metabolic pathway. Initial SAR results showed that this metabolic pathway could be disfavored by shortening of the N-alkyl side chain as shown with some benzyl derivatives (COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound15, 19–20 and 22–23).
After this initial exploration, compounds 19 and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound20 were identified which combined good potency and metabolic stability. Compound COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound20 was further explored for its ability to potentiate the in vitro concentration response curve (CRC) of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundglutamate on cloned human mGluR2.
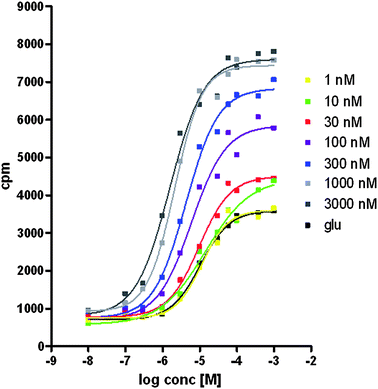
Results indicative of positive allosteric modulation were observed. As shown in Fig. 3, the CRC of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundglutamate shifts to the left and upwards with increasing concentrations of compound COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound20. A 6.6-fold shift in the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundglutamate EC50 was seen in the presence of 3 μM of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound20(glutamate pEC50 was 4.9 and 5.8 in the absence or presence of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound20, respectively).
Conclusions
In summary, a novel series of 5-substituted isoquinolones with mGluR2 PAM activity has been identified. Initial SAR studies from the HTS singleton 7 confirmed the COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundisoquinolone series 6 as a novel and viable mGluR2 PAM chemotype. Preliminary SAR data suggest that (a) lipophilicity on the distal phenyl ring seems to be good for activity (b) a COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundhydrogen bond donor at position C-5 is not required for activity, (c) the distance between the core and the phenyl ring is important, with a 3 COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcarbon spacer optimal for activity. At present, metabolic stability is the main issue identified for this novel chemotype, nevertheless the shorter benzyl derivatives may allow to circumvent this issue. From our first round exploration, we identified compounds (19 and COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound20) with good potency in the GTPγS assay and improved metabolic stability in human liver microsomes. Compound COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound20 proved to be a positive allosteric modulator of mGluR2 by its ability to potentiate the in vitro CRC of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundglutamate, deserving further optimization. Further evaluation of this compound as well as a broader SAR exploration are underway and will be reported in due course.
Experimental
Biology
Membrane preparation.
CHO-cells were cultured to pre-confluence and stimulated with 5 mM COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundbutyrate for 24 h, prior to washing in PBS, and then collection by scraping in homogenisationbuffer (50 mM Tris-HCl buffer, pH 7.4, 4 °C). Cell lysates were homogenized briefly (15 s) using an ultra-turrax homogenizer. The homogenate was centrifuged at 23 500 × g for 10 min and the supernatant discarded. The pellet was resuspended in 5 mM Tris-HCl, pH 7.4 and centrifuged again (30 000 × g, 20 min, 4 °C). The final pellet was resuspended in 50 mM HEPES, pH 7.4 and stored at −80 °C in appropriate aliquots before use. Protein concentration was determined by the Bradford method (Bio-Rad, USA) with bovine serum albumin as standard.
[35S]GTPγS binding assay.
Measurement of mGluR2 positive allosteric modulatory activity of test compounds in membranes containing human mGluR2 was performed using frozen membranes that were thawed and briefly homogenised prior to pre-incubation in 96-well microplates (15 μg/assay well, 30 min, 30 °C) in assay buffer (50 mM HEPES pH 7.4, 100 mM COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundNaCl, 3 mM COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMgCl2, 50 μM GDP, 10 μg ml−1saponin) with increasing concentrations of positive allosteric modulator (from 0.3 nM to 50 μM) and either a minimal pre-determined concentration of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundglutamate (PAM assay), or no added COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundglutamate. For the PAM assay, membranes were pre-incubated with COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundglutamate at EC25 concentration, i.e. a concentration that gives 25% of the maximal response COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundglutamate. After addition of [35S]GTPγS (0.1 nM, f.c.) to achieve a total reaction volume of 200 μl, microplates were shaken briefly and further incubated to allow [35S]GTPγS incorporation on activation (30 min, 30 °C). The reaction was stopped by rapid vacuum filtration over glass-fibre filter plates (Unifilter 96-well GF/B filter plates, Perkin-Elmer, Downers Grove, USA) microplate using a 96-well plate cell harvester (Filtermate, Perkin-Elmer, USA), and then by washing three times with 300 μl of ice-cold wash buffer (Na2PO4·2H2O 10 mM, NaH2PO4·H2O 10 mM, pH = 7.4). Filters were then air-dried, and 40 μl of liquid scintillation cocktail (Microscint-O) was added to each well, and membrane-bound [35S]GTPγS was measured in a 96-well scintillation plate reader (Top-Count, Perkin-Elmer, USA). Non-specific [35S]GTPγS binding is determined in the presence of cold 10 μM COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundGTP. Each curve was performed at least three times using duplicate sample per data point and at 11 concentrations.
General procedures
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound5-(4-Methoxyphenethylamino)-2-propylisoquinolin-1(2H)-one (COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound10).
To a solution of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound5-chloroisoquinolin-1(2H)-one24 (596 mg, 2.66 mmol) in COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundDMF (10 mL) at 0 °C was added COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundsodium hydride (67 mg, 2.8 mmol) and the mixture was stirred for 15 min. COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound1-Bromopropane (0.27 mL, 2.93 mmol) was added and the mixture was stirred at room temperature for 12 h. The reaction mixture was poured into COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundwater and extracted with COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundEtOAc (2 × 25 mL). The combined organic layers were dried (MgSO4), filtered and concentrated in vacuo. The residue was purified flash chromatography (AIT Flashsmart prepacked column 25 g SiO2, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCH2Cl2) to afford COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound25: orange solid, yield 396. mg (56%). 1H- NMR: (300MHz, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCDCl3) δ 1.00 (t, J = 7.4 Hz, 3H), 1.80 (m, 2H), 3.91 (t, J = 7.2 Hz, 2H), 6.82 (d, J = 7.7 Hz, 1H), 7.10 (d, J = 7.7 Hz, 1H), 7.29 (d, J = 7.7 Hz, 1H), 7.81 (dd, J = 7.7 Hz, 1H), 8.33 (d, J = 7.7 Hz, 1H). ESI-MS: m/z 266.9 [M + H]+.
To a mixture of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound5-bromo-2-propylisoquinolin-1(2H)-oneCOMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound25 (287 mg, 1.08 mmol), t-BuONa (160 mg, 1.62 mmol), Pd2(dba)3 (50 mg, 0.054 mmol), COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundBINAP (34 mg, 0.054 mmol) in degassed dry COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundtoluene (4 mL) was added 4-methoxyphenetyl amine (250 mg, 1.62 mmol). The reaction mixture was heated (sealed tube) at 180 °C for 1 h under microwave irradiation. The mixture was cooled to room temperature and filtered through Celite®. The reaction mixture was poured into COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundwater and extracted with COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCH2Cl2 (2 × 25 mL). The combined organic layers were dried (Na2SO4), filtered and concentrated in vacuo. The crude product was purified by flash chromatography (AIT Flashsmart prepacked column 25 g, SiO2, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundcyclohexane/AcOEt 90/10) to afford COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound10: white solid. mp.: 110 °C. 1H- NMR: (300MHz, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCDCl3) δ 0.96 (t, J = 7.2 Hz, 3H), 1.02 (t, J = 7.2 Hz, 2H), 1.80 (m, 2H), 2.97 (d, J = 6.9 Hz, 2H), 3.43–3.49 (m, 3H), 3.80 (s, 3H), 3.94 (t, J = 7.5 Hz, 2H), 6.26 (d, J = 7.8 Hz, 1H), 6.87 (d, J = 8.7 Hz, 3H), 7.00 (d, J = 7.5 Hz, 1H), 7.17 (d, J = 8.7 Hz, 2H), 7.36 (t, J = 8.1 Hz, 1H), 7.85 (d, J = 8.1 Hz, 1H). ESI-MS: m/z 337.4 [M + H]+.
The procedure used for compound COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound10 was further used to prepare the following compounds:
COMPOUND LINKS
Read more about this on ChemSpider
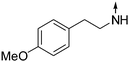
Download mol file of compound5-Phenethylamino-2-propylisoquinolin-1(2H)-one (7).
Obtained from COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundphenethylamine. 7: orange solid. mp.: 120 °C. 1H-NMR: (300MHz, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCDCl3) δ 0.96 (t, J = 7.2 Hz, 3H), 1.79 (m, 2H), 3.05 (t, J = 6.9 Hz, 2H), 3.51 (t, J = 6.9 Hz, 2H), 3.95 (t, J = 7.2 Hz, 2H), 6.32 (m, 1H), 7.02 (d, J = 7.5 Hz, 1H), 7.23–7.27 (m, 3H), 7.36 (m, 4H), 7.90 (m, 1H). ESI-MS: m/z 307.3 [M + H]+.
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound5-(2-Methoxyphenethylamino)-2-propylisoquinolin-1(2H)-one (COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound8).
Obtained from COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound2-methoxyphenethylamine. COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound8: off-white solid. mp.: 98 °C. 1H-NMR: (300MHz, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCDCl3) δ 0.96 (t, J = 7.4 Hz, 3H), 1.8 (m, 2H), 3.05 (t, J = 6.7 Hz, 2H), 3.4 (t, J = 6.7 Hz, 2H), 3.9 (t, J = 7.4 Hz, 2H), 4.0 (t, J = 7.4 Hz, 2H), 6.3 (d, J = 7.8 Hz, 1H), 6.92 (d, J = 8.4 Hz, 2H), 6.95 (td, J = 7.4 Hz, J = 1.0 Hz, 1H), 7.02 (d, J = 7.8 Hz, 1H), 7.2 (dd, J = 7.4 Hz, J = 1.8 Hz, 1H), 7.25 (dd, J = 7.4 Hz, J = 1.8 Hz, 1H), 7.3 (d, 1H), 7.38 (t, J = 7.8 Hz, 1H), 7.8 (d, J = 8.2 Hz, 1H). ESI-MS: m/z 337.3 [M + H]+.
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound5-(3-Methoxyphenethylamino)-2-propylisoquinolin-1(2H)-one (9).
Obtained from COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound3-methoxyphenethylamine. 9: white solid. mp.: 124 °C 1H-NMR: (300MHz, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCDCl3) δ 0.98 (t, J = 7.2 Hz, 3H), 1.82 (m, 2H), 3.02 (t, J = 7.0 Hz, 2H), 3.51 (t, J = 6 Hz, 2H), 3.81 (s, 3H), 3.97 (t, J = 7.8 Hz, 2H), 6.27 (d, J = 7.8 Hz, 1H), 6.85 (m, 4H), 7.02 (t, J = 7.4 Hz, 1H), 7.28 (m, 1H), 7.38 (t, J = 8.4 Hz, 1H), 7.85 (d, J = 8.4 Hz, 1H). ESI-MS: m/z 337.4 [M + H]+.
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound5-(4-Methylphenethylamino)-2-propylisoquinolin-1(2H)-one (11).
Obtained from COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound2-methoxyphenethylamine. 11: white solid, mp.: 127 °C. 1H- NMR: (300MHz, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCDCl3) δ 0.96 (t, J = 7.4 Hz, 3H), 1.81 (m, 2H), 2.20 (s, 3H), 2.97 (t, J = 6.8 Hz, 2H), 3.47 (t, J = 6.8 Hz, 2H), 3.97 (t, J = 7.4 Hz, 2H), 6.26 (d, J = 7.2 Hz, 1H), 6.82 (d, J = 7.2 Hz, 1H), 6.92 (d, J = 7.6 Hz, 1H), 7.05 (m, 3H), 7.15 (s, 1H), 7.31 (t, J = 7.9 Hz, 1H), 7.80 (d, J = 7.9 Hz, 1H). ESI-MS: m/z 320.4 [M + H]+.
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound5-[N-(4-Methoxyphenethyl)-N-methylamino]-2-propylisoquinolin-1(2H)-one (COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound12).
Obtained from COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundN-(4-Methoxyphenethyl)-N-methylamine. COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound12: yellow oil. 1H-NMR: (300MHz, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCDCl3) δ 1.00 (t, J = 7.4 Hz, 3H), 1.80 (m, 2H), 2.83 (t, J = 7.4 Hz, 2H), 2.87 (s, 3H), 3.25 (t, J = 7.6 Hz, 2H), 3.81 (s, 3H), 4.01 (t, J = 7.4 Hz, 2H), 6.71 (d, J = 7.6 Hz, 2H), 6.80 (dd, J = 6.5 Hz, 2H), 7.01 (d, J = 7.6 Hz, 1H), 7.10 (d, J = 8.7 Hz, 2H), 7.35 (d, J = 7.4 Hz, 1H), 7.43 (t, J = 8.0 Hz, 1H), 8.16 (d, J = 8.0 Hz, 1H). ESI-MS: m/z 350.5 [M + H]+.
COMPOUND LINKS
Read more about this on ChemSpider
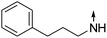
Download mol file of compound5-(3-Phenylpropylamino)-2-propylisoquinolin-1(2H)-one (COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound14).
Obtained from COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound3-phenylpropylamine. COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound14: white solid. mp.:118 °C. 1H-NMR: (300MHz, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCDCl3) δ 0.97 (t, J = 7.5 Hz, 3H), 1.80 (s, J = 7.5 Hz, 2H), 2.08 (q, J = 7.2 Hz, 2H), 2.79 (t, J = 7.5 Hz, 2H), 3.26 (t, J = 7.2 Hz, 2H), 3.95 (t, J = 7.2 Hz, 2H), 6.29 (d, J = 7.5 Hz, 1H), 6.83 (m, 1H), 7.02 (d, J = 7.8 Hz, 1H), 7.28 (m, 7H), 7.85 (d, J = 8.1 Hz, 1H). ESI-MS: m/z 320.3 [M + H]+.
COMPOUND LINKS
Read more about this on ChemSpider
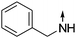
Download mol file of compound5-Benzylamino-2-propylisoquinolin-1(2H)-one (COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound15).
Obtained from COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundbenzylamine. COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound15: orange solid. mp.: 111 °C. 1H-NMR: (300MHz, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCDCl3) δ 0.97 (t, J = 7.2 Hz, 3H), 1.81 (m, 2H), 3.96 (t, J = 7.2 Hz, 2H), 4.44 (s, 2H), 7.05 (d, J = 7.5 Hz, 1H), 6.49 (m, 1H), 7.36 (m, 7H), 7.91 (m, 1H). ESI-MS: m/z 293.4 [M + H]+.
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound5-(3-Methoxybenzylamino)-2-propylisoquinolin-1(2H)-one (COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound16).
Obtained from COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound3-methoxybenzylamine. COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound16: yellow solid. mp.: 140 °C. 1H-NMR: (300MHz, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCDCl3) δ 0.99 (t, J = 7.2 Hz, 3H), 1.61 (s, 2H), 1.83 (m, 2H), 3.83 (s, 3H), 3.98 (t, J = 7.2 Hz, 2H), 4.42 (s, 3H), 6.45 (d, J = 7.9 Hz, 1H), 6.85 (m, 2H), 7.07 (m, 3H), 7.35 (m, J = 7.9 Hz, 2H), 7.87 (d, J = 4.0 Hz, 1H). ESI-MS: m/z 323.4 [M + H]+.
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound5-(4-Methoxybenzylamino)-2-propylisoquinolin-1(2H)-one (COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound17).
Obtained from COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound4-methoxybenzylamine. COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound17: white solid. mp.:140 °C. 1H-NMR: (300MHz, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCDCl3) δ 0.95 (t, J = 7.2 Hz, 3H), 1.81 (m, 2H), 3.80 (s, 3H), 3.98 (t, J = 7.4 Hz, 2H), 4.41 (s, 2H), 6.88 (m, J = 6.7 Hz, 2H), 7.08 (m, J = 7.4 Hz, 1H), 7.31 (m, 3H), 7.35 (m, J = 7.9 Hz, 3H). ESI-MS: m/z 323.4 [M + H] +.
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound5-(3-Phenoxybenzylamino)-2-propylisoquinolin-1(2H)-one (COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound18).
Obtained from COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound3-phenoxybenzylamine. COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound18: beige solid. mp.: 128 °C. 1H-NMR: (300MHz, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCDCl3) δ 1.00 (t, J = 7.4 Hz, 3H), 1.80 (q, J = 7.4 Hz, 2H), 3.99 (t, J = 8.4 Hz, 2H), 4.41 (s, 2H), 6.46 (d, J = 7.7 Hz, 1H), 6.77 (d, J = 7.7 Hz, 1H), 7.05 (m, 7H), 7.28 (m, 4H), 7.86 (d, J = 7.7 Hz, 1H). ESI-MS: m/z 385.5 [M + H]+.
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound2-Propyl-5-(3-trifluoromethoxybenzylamino)isoquinolin-1(2H)-one (19).
Obtained from COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound3-trifluoromethoxybenzylamine. 19: beige solid. 1H-NMR: (300MHz, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCDCl3) δ 0.98 (t, J = 7.4 Hz, 3H), 1.80 (q, J = 7.4 Hz, 2H), 3.96 (t, J = 7.4 Hz, 2H), 4.55 (s, 2H), 6.90 (d, J = 7.4 Hz, 1H), 7.22 (m, 6H), 7.60 (m, 1H), 8.45 (d, J = 7.2 Hz, 1H). ESI-MS: m/z 377.4 [M + H]+.
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound5-(2-Chlorobenzylamino)-2-propylisoquinolin-1(2H)-one (COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound20).
Obtained from COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound2-chlorobenzylamine. COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound20: light yellow. mp.:140 °C. 1H-NMR: (300MHz, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCDCl3) δ 0.98 (t, J = 7.4 Hz, 3H), 1.81 (q, J = 7.4 Hz, 2H), 3.99 (t, J = 8.4 Hz, 2H), 4.55 (s, 2H), 6.46 (d, J = 7.7 Hz, 1H), 6.77 (d, J = 7.7 Hz, 1H), 7.08 (m, 1H), 7.29 (m, 3H), 7.42 (m, 2H), 7.86 (d, J = 8.2 Hz, 1H). ESI -MS: m/z 327.8 [M + H]+.
5-(4-Methoxyphenetoxy)2-propylisoquinolin-1(2H)-one (13).
To a solution of 5-(4-methoxyphenetoxy)isoquinolin-1(2H)-one28 (100 mg, 0.34 mmol) in COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundDMF (10 mL) at 0 °C was added COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundsodium hydride (10 mg, 0.41 mmol) and the mixture was stirred for 15 min. COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound1-Bromopropane (0.035 mL, 0.38 mmol) was added The mixture was heated (sealed tube) at 180 °C for 15 min under microwave irradiation. The mixture was cooled to room temperature and filtered through Celite®. The reaction mixture was poured into COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundwater and extracted with COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundEtOAc (2 × 25 mL). The combined organic layers were dried (MgSO4), filtered and concentrated in vacuo. The residue was purified by flash chromatography (AIT Flashsmart prepacked column 25 g SiO2, 2% COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundMeOH in COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCH2Cl2) to afford 13: orange oil, yield 20. mg (17%). 1H-NMR: (300MHz, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCDCl3) δ 0.97 (t, J = 7.5 Hz, 3H), 1.81 (m, 2H), 3.13 (t, J = 6.3 Hz, 2H), 3.80 (s, 3H), 3.96 (t, J = 7.5 Hz, 2H), 4.24 (t, J = 7.2 Hz, 2H), 6.85 (m, 3H), 7.02 (m, 2H), 7.24 (d, J = 8.7 Hz, 2H), 7.36 (t, J = 8.4 Hz, 1H), 7.99 (d, J = 8.1 Hz, 1H). ESI-MS: m/z 338 [M + H]+.
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound5-Bromo-8-chloro-2-propylisoquinolin-1(2H)-one (COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound32).
To a solution of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound3-bromo-6-chloro-2-methyl-N-propylbenzamideCOMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound31 (1.77 g, 6.09 mmol) in dry COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundTHF (100 mL) at −78 °C and under nitrogen atmosphere, was added a 1.8M solution of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundlithium diisopropylamide (8.46 mL, 15.2 mmol) over 20 min. The reaction mixture was stirred at −78 °C for 2 h, then dry COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundDMF (2.35 mL, 30.5 mmol) was added dropwise over 10 min. After 10 more minutes, the reaction mixture was allowed to warm up untill −10 °C and was hydrolysed slowly with a 6.0 N solution of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundHCl. The reaction mixture was extracted with AcOEt (3 × 25 mL), dried over MgSO4, filtered and concentrated in vacuo. The residue was triturated in COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compounddiisopropylether, filtered and dried, to afford COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound32: brown solid, yield 1.3 g (71%). ESI-MS: m/z 300–304 [M + H]+.
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound8-Chloro-5-(phenethylamino)-2-propylisoquinolin-1(2H)-one (COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound21).
To a mixture of COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound5-bromo-8-chloro-2-propylisoquinolin-1(2H)-oneCOMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound32 (200 mg, 0.665 mmol), t-BuONa (96 mg, 0.998 mmol), Pd2(dba)3 (31 mg, 0.033 mmol), COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundBINAP (41 mg, 0.067 mmol) in degassed dry COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundtoluene (7 mL) was added COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundphenethylamine (81 mg, 0.665 mmol). The reaction mixture was heated at 110 °C for 2 h. The mixture was cooled to room temperature and filtered through Celite®. The reaction mixture was poured into COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundwater and extracted with COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundEtOAc (2 × 10 mL). The combined organic layers were dried (Na2SO4), filtered and concentrated in vacuo. The crude product was purified by flash chromatography (AIT Flashsmart prepacked column 15 g, SiO2, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundDCM–AcOEt 98![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) to afford COMPOUND LINKS
2) to afford COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound21: beige solid. mp.: 107 °C. 1H-NMR: (300MHz, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCDCl3) δ 0.96 (t, J = 7.4 Hz, 3H), 1.78 (m, 2H), 2.99 (d, J = 7.3 Hz, 2H), 3.45 (t, J = 7.4 Hz, 3H), 3.80 (t, J = 7.4 Hz, 3H), 6.26 (s, 1H), 6.87–7.42 (m, 8H). ESI-MS: m/z 341–343 [M + H]+.
The procedure used for compound COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound21 was further used to prepare the following compounds:
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound5-(Benzylamino)-8-chloro-2-propylisoquinolin-1(2H)-one (COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound22).
Obtained from COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundbenzylamine. COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound22: brown solid. mp.: 152 °C. 1H-NMR: (300MHz, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCDCl3) δ 0.95 (t, J = 7.4 Hz, 3H), 1.73 (m, 2H), 3.87 (t, J = 7.4 Hz, 2H), 4.38 (s, 2H), 6.34 (s, 1H), 6.88 (m, 1H), 7.02 (m, 1H), 7.23 (m, 6H). ESI-MS: m/z 327–329 [M + H]+.
COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound8-Chloro-5-(3-methoxybenzylamino)-2-propylisoquinolin-1(2H)-one (COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound23).
Obtained from COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound(3-methoxyphenyl)methanamine. COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compound23: white solid. mp.: 152 °C. 1H-NMR: (300MHz, COMPOUND LINKS
Read more about this on ChemSpider
Download mol file of compoundCDCl3) δ 0.98 (t, J = 7.4 Hz, 3H), 1.82 (q, J = 7.4 Hz, 2H), 3.89 (s, 3H), 3.94 (t, J = 7.7 Hz, 2H), 4.41 (s, 2H), 6.34 (d, J = 7.7 Hz, 1H), 6.84 (d, J = 8.4 Hz, 1H), 6.86–6.98 (m, 4H), 7.05 (m, 1H). ESI-MS: m/z 357–359 [M + H]+.
Notes and references
- C. J. Swanson, M. Bures, M. P. Johnson, A. M. Linden, J. A. Monn and D. D. Schoepp, Nat. Rev. Drug Discovery, 2005, 4, 131–144 CrossRef CAS.
- D. D. Schoepp and G. J. Marek, Curr. Drug Targets: CNS Neurol. Disord., 2002, 1, 215–225 Search PubMed.
- P. J. Conn, C. W. Lindsley and C. K. Jones, Trends Pharmacol. Sci., 2009, 30, 25–31 CrossRef CAS.
- L. Levine, B. Gaydos, D. Sheehan, A. Goddard, J. Feighner, W. Potter and D. D. Schoepp, Neuropharmacology, 2002, 43, 294–295.
- D. D. Schoepp, R. A. Wright, L. R. Levine and B. Gaydos, Stress, 2003, 6, 189–197 CrossRef CAS.
- S. T. Patil, L. Zhang, F. Martenyi, S. L. Lowe, K. A. Jackson, B. V. Andreev, A. S. Avedisova, L. M. Bardenstein, I. Y. Gurovich, M. A. Morozova, S. N. Mosolov, N. G. Neznanov, A. M. Reznik, A. B. Smulevich, V. A. Tochilov, B. G. Johnson, J. A. Monn and D. D. Schoepp, Nat. Med., 2007, 13, 1102–1107 CrossRef CAS.
- M. J. Fell, K. A. Svensson, B. G. Johnson and D. D. Schoepp, J. Pharmacol. Exp. Ther., 2008, 326, 209–217 CrossRef CAS.
- J. P. Conn, A. Christopoulos and C. W. Lindsley, Nat. Rev. Drug Discovery, 2009, 8, 41–54 CrossRef CAS.
-
(a) M. E. Farley, Expert Opin. Ther. Pat., 2009, 19, 1259–1275 Search PubMed;
(b) A. A. Trabanco, J. M. Cid, H. Lavreysen, G. J. Macdonald and G. Tresadern, Curr. Med. Chem., 2011 Search PubMed , in press.
- E. J. Brnardic, M. E. Fraley, R. M. Garbaccio, M. E. Layton, J. M. Sanders, C. Culberson, M. A. Jacobson, B. C. Magliaro, P. H. Hutson, J. A. O'Brien, S. L. Huszar, J. M. Uslaner, K. L. Fillgrove, C. Tang, Y. Kuo, M. S. Sylvie and G. D. Hartman, Bioorg. Med. Chem. Lett., 2010, 20, 3129–3133 CrossRef CAS.
- G. Tresadern, J. M. Cid, G. J. Macdonald, J. A. Vega, A. I. de Lucas, A. García, E. Matesanz, M. L. Linares, D. Oehlrich, H. Lavreysen, I. Biesmans and A. A. Trabanco, Bioorg. Med. Chem. Lett., 2010, 20, 175–179 CrossRef CAS.
- R. M. Garbaccio, E. J. Brnardic, M. E. Farley, G. D. Hartman, P. H. Hutson, A. J. O'Brien, B. C. Magliaro, J. M. Uslaner, S. L. Huszar, K. L. Fillgrove, J. H. Small, C. Tang, Y. Kuo and M. A. Jacobson, ACS Med. Chem. Lett., 2010, 1, 406–410 Search PubMed.
- The effect of these compounds on the [35S]-GTPγS binding induced by 4 μM glutamate (∼EC20) was characterized using a CHO cell line expressing the human mGluR2 receptor.
- J. Roth, F. Madoux, P. Hodder and W. R. Roush, Bioorg. Med. Chem. Lett., 2008, 18, 2628–2632 CrossRef CAS.
- M. M. Robison and B. L. Robison, J. Org. Chem., 1956, 21, 1337–1341 CrossRef CAS.
-
H. Kodama, T. Katsuhira, T. Nishida, T. Hino and T. K. Tomokazu, WO2001083421 A1, 2001.
- P. Beak, S. T. Kerrick and D. J. Gallagher, J. Am. Chem. Soc., 1993, 115, 10628–10636 CrossRef CAS.
- For further details on these two compounds see Reference compounds BINA and 2,2,2-TEMPS were tested in house in the GTPγS assay for comparison, showing pEC50 = 7.12 (EMAX = 213%) and pEC50 = 7.5 (EMAX = 123%) respectively. ref. 9a and 9b.
|
| This journal is © The Royal Society of Chemistry 2011 |












![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20) to afford 31: yellow solid, yield 10.5 g (90%). ESI-MS: m/z 290–294 [M + H]+.
20) to afford 31: yellow solid, yield 10.5 g (90%). ESI-MS: m/z 290–294 [M + H]+.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) to afford 21: beige solid. mp.: 107 °C. 1H-NMR: (300MHz, CDCl3) δ 0.96 (t, J = 7.4 Hz, 3H), 1.78 (m, 2H), 2.99 (d, J = 7.3 Hz, 2H), 3.45 (t, J = 7.4 Hz, 3H), 3.80 (t, J = 7.4 Hz, 3H), 6.26 (s, 1H), 6.87–7.42 (m, 8H). ESI-MS: m/z 341–343 [M + H]+.
2) to afford 21: beige solid. mp.: 107 °C. 1H-NMR: (300MHz, CDCl3) δ 0.96 (t, J = 7.4 Hz, 3H), 1.78 (m, 2H), 2.99 (d, J = 7.3 Hz, 2H), 3.45 (t, J = 7.4 Hz, 3H), 3.80 (t, J = 7.4 Hz, 3H), 6.26 (s, 1H), 6.87–7.42 (m, 8H). ESI-MS: m/z 341–343 [M + H]+.
