 Open Access Article
Open Access ArticleThe RNA–DNA hybrid structure determined by EPR, CD and RNase H1†
Olga
Romainczyk
a,
Burkhard
Endeward
b,
Thomas F.
Prisner
b and
Joachim W.
Engels
*a
aInstitute of Organic Chemistry and Chemical Biology, Max-von-Laue-Str. 7, 60438 Frankfurt am Main, Germany. E-mail: Joachim.engels@chemie.uni-frankfurt.de; Fax: +49 69-79829148; Tel: +49 69-79829150
bInstitute of Physical and Theoretical Chemistry and Center of Biomolecular Magnetic Resonance, Max-von-Laue-Str. 7, 60438 Frankfurt am Main, Germany. E-mail: prisner@prisner.de
First published on 19th February 2011
Abstract
A or B: RNA–DNA hybrids, key intermediates in gene regulation, were classified by pulsed electron–electron double resonance (PELDOR) in combination with CD spectroscopy into two classes, interpreted as A- and B-like structures. RNase H1 cleavage of these hybrids is in full agreement with these assignments, cleaving the hybrids with A-like geometry preferentially. This combined analytical approach allows the interpretation and eventually the design of more easily cleavable hybrids as needed for the antisense technology.
RNA–DNA hybrid helices, first reported by Rich 50 years ago, are important intermediates in biology and play an essential role in transcription of DNA into RNA, replication of double-stranded DNA and reverse transcription of viral RNA into DNA.1 They are also key to be recognized by degrading enzymes like ribonuclease H (RNase H). The latter enzyme is an essential component of the antisense technique that degrades mRNA and reduces gene expression.2,3 Understanding the molecular mechanism of these processes depends on more detailed knowledge of the structure of the hetero duplexes.
Electron Paramagnetic Resonance (EPR), in particular pulsed electron–electron double resonance (PELDOR),4 can be applied to study long range distance constraints up to 7 nm of oligonucleotides.5 The accuracy of the method is high enough to distinguish between B and A-helix forms6,7 as present in DNA and RNA or for DNA in the presence of trifluoroethanol.8 We successfully established the organic nitroxide TPA (2,2,5,5-tetramethyl-3-pyrrolin-N-oxyl-3-acetylene, Fig. 1) as a rigid paramagnetic label for DNA and RNA.9 In addition by adding the label at a defined position on the heterocyclic base moiety we can direct it to either the major or minor groove of the double helix. Adding the spin label at the 5-position of pyrimidines it points towards the major groove, whereas at the 2-position of purines minor grooves are addressed.
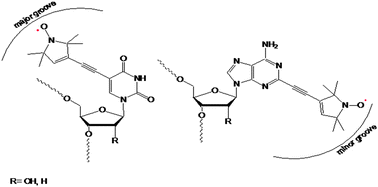 | ||
Fig. 1 Left: r![[U with combining low line]](https://www.rsc.org/images/entities/b_char_0055_0332.gif) /d /d![[T with combining low line]](https://www.rsc.org/images/entities/b_char_0054_0332.gif) labelled on 5-position with TPA; right: r labelled on 5-position with TPA; right: r![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) /d /d![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) labelled on 2-position with TPA. labelled on 2-position with TPA. | ||
We chose non-self-complementary 15-mer sequences with identical base sequence for DNA and RNA except for T and U.
• DNA(d![[T with combining low line]](https://www.rsc.org/images/entities/b_char_0054_0332.gif) ) 5′-CGCTACATAG
) 5′-CGCTACATAG![[T with combining low line]](https://www.rsc.org/images/entities/b_char_0054_0332.gif) GAGC-3′
GAGC-3′
• DNA(d![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) ) 5′-GCTCACTATGT
) 5′-GCTCACTATGT![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) GCG-3′
GCG-3′
• RNA(r![[U with combining low line]](https://www.rsc.org/images/entities/b_char_0055_0332.gif) ) 5′-CGCUACAUAG
) 5′-CGCUACAUAG![[U with combining low line]](https://www.rsc.org/images/entities/b_char_0055_0332.gif) GAGC-3′
GAGC-3′
• RNA(r![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) ) 5′-GCUCACUAUGU
) 5′-GCUCACUAUGU![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) GCG-3′
GCG-3′
where the underlined bases carry the spin label. The spin label was placed in the more rigid center part, at least 3 bases apart from either end. Based on PyMol modeling the geometry of the angles and distances should show optimal discrimination between A- (2.5 nm) and B-form (2.9 nm) when hybridized. In our case an optimal distance appeared to be between 2 and 4 nm and the orientation of the two spin labels to each other was similar. From the 8 different duplexes resulting 6 were chosen: DNA(d![[T with combining low line]](https://www.rsc.org/images/entities/b_char_0054_0332.gif) )/DNA(d
)/DNA(d![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) ) to RNA(r
) to RNA(r![[U with combining low line]](https://www.rsc.org/images/entities/b_char_0055_0332.gif) )/RNA(r
)/RNA(r![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) )via the two possible hybrids DNA(d
)via the two possible hybrids DNA(d![[T with combining low line]](https://www.rsc.org/images/entities/b_char_0054_0332.gif) )/RNA(r
)/RNA(r![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) ), DNA(d
), DNA(d![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) )/RNA(r
)/RNA(r![[U with combining low line]](https://www.rsc.org/images/entities/b_char_0055_0332.gif) ) with spin label and two without DNA(dT)/RNA(rA) and DNA(dA)/RNA(rU).
) with spin label and two without DNA(dT)/RNA(rA) and DNA(dA)/RNA(rU).
RNA(rU) and (rA) were synthesized via “on column” solid phase synthesis as reported earlier.7 For DNA synthesis the 2-iodo-deoxy-adenosine building block had to be prepared. This we accomplished via an analogous synthesis to the RNA building block but had to change the protecting groups in order to get better yields using milder conditions (see ESI†). The DNA oligomers were synthesized by phosphoramidite chemistry on a 1 μmol scale, with the same coupling time for all used phosphoramidites. The RNAs were synthesized on a 0.2 μmol scale with phosphoramidites purchased from Dharmacon (ACE chemistry).10
All oligonucleotides were synthesized completely without interrupting the synthesis after incorporation of the iodinated phosphoramidite. For the Sonogashira cross coupling the column was removed from the synthesizer, maintained under an argon atmosphere and acetylenic TPA was added. To achieve better yields, the Sonogashira cross coupling was performed two times with the same amount of reagents.
Finally, the DNA was cleaved from the support and completely deprotected with ammonia. In the case of RNA, the cleavage of methyl-groups of the phosphate was done first using disodium-2-carbamoyl-2-cyanoethylene-1,1-dithiolate-trihydrate. Afterwards, in one step the oligonucleotide was cleaved from the solid-support (polystyrene) and the functional groups were deprotected with methylamine and purified by anion-exchange HPLC with 1 M LiCl. Prior to use for ACE chemistry deprotection of the 2′-hydroxy group is performed by TEMED/acetic acid buffer, pH 3.8. The DNA and RNA synthesised were characterized by electron spray ionization (ESI) spectra (Table 1, ESI†). UV melting curves (Tm) of the duplexes dissolved in phosphate buffer (10 mM Na2HPO4, 10 mM NaH2PO4, 140 mM NaCl, 0.01 mM duplex, pH 7) were recorded on a UV-/VIS-600 from 5 to 95 °C with a heating rate of 0.5 °C min−1. CD spectra were measured on a JASCO J-710 spectropolarimeter with a Peltier thermostat (Table 1). The duplexes for the distance measurements were dissolved in the same buffer with addition of 20% ethylene glycol and at the 10× concentration as for the Tm-measurements.
| Duplex | T m a/°C | Distance rb/nm | λ 1 c/nm | t 1/2 d/min | Helix type |
|---|---|---|---|---|---|
| a In phosphate buffer, pH 7. b Measured by PELDOR with addition of 20% ethylene glycol. c The maximum in CD spectra to differentiate the helix type. d Treatment with RNase H at 37 °C, calculated by extrapolation of the data: [/], not spin labelled; [—], no degradation. | |||||
DNA(d![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) )/RNA(r )/RNA(r![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) ) ) |
47.8 ± 0.2 | 2.8 ± 0.1 | 270 | 25.8 ± 4.6 | A-like form |
DNA(d![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) )/RNA(r )/RNA(r![[U with combining low line]](https://www.rsc.org/images/entities/b_char_0055_0332.gif) ) ) |
45.7 ± 0.2 | 3.0 ± 0.05 | 282 | 147.8 ± 55.9 | B-like form |
DNA(d![[T with combining low line]](https://www.rsc.org/images/entities/b_char_0054_0332.gif) )/DNA(d )/DNA(d![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) ) ) |
48.6 ± 0.1 | 3.0 ± 0.05 | 276 | [—] | B-form |
RNA(r![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) )/RNA(r )/RNA(r![[U with combining low line]](https://www.rsc.org/images/entities/b_char_0055_0332.gif) ) ) |
63.5 ± 0.4 | 2.7 ± 0.1 | 266 | [—] | A-form |
| DNA(dT)/RNA(rA) | 50.9 ± 0.2 | [/] | 273 | 110.9 ± 11.5 | B-like form |
| DNA(dA)/RNA(rU) | 49.4 ± 0.2 | [/] | 277 | 130.9 ± 40.9 | B-like form |
The dead-time free 4-pulse PELDOR sequence was used for all experiments.11 X-band PELDOR data were collected at 40 K with a detection window of 2.5 μs. Further experimental details are given in the ESI.† The distance measured12 for the modified RNA double helix was 2.7 nm and for the DNA double helix 3.0 nm (see Fig. 2) which nicely fits the expected distances for an A- (2.5 nm) and B-helix (2.9 nm) geometry. For the hybrids we unexpectedly obtained two different distances, depending on the labelling position. When hybridizing labelled DNA(d![[T with combining low line]](https://www.rsc.org/images/entities/b_char_0054_0332.gif) ) with labelled RNA(r
) with labelled RNA(r![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) ) we measured 2.8 nm but with labelled DNA(d
) we measured 2.8 nm but with labelled DNA(d![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) ) and labelled RNA(r
) and labelled RNA(r![[U with combining low line]](https://www.rsc.org/images/entities/b_char_0055_0332.gif) ) we obtained a distance of 3.0 nm for the mixed hybrid (Table 1). Therefore the first hybrid is close to the distance measured for RNA, whereas the second is identical to the distance measured for DNA.
) we obtained a distance of 3.0 nm for the mixed hybrid (Table 1). Therefore the first hybrid is close to the distance measured for RNA, whereas the second is identical to the distance measured for DNA.
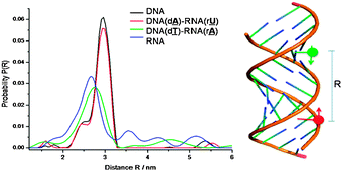 | ||
| Fig. 2 PELDOR determined distances distribution function P(R) between the two TPA spin labels of duplexes (DNA, DNA/RNA and RNA). | ||
To our surprise these two classes of spectra are also reflected in the UV and CD spectra of the spin labelled duplexes as well as in the reduced hyperchromicity during melting as observed by UV for DNA like structures compared to RNA (see Fig. 3 and detailed CD analysis in ESI†). Therefore the distances between the spin labels measured (B- to A-like) can be grouped DNA/DNA ≈ DNA(dA)–RNA(rU) > DNA(dT)–RNA(rA) ≈ RNA/RNA. The only difference between the two hybrids is the attachment side for the spin label. Here the spin label at the U5 position points towards the major groove whereas the label at the 2-position on A is directed to the open minor groove (Fig. 1). Considering the differences between the major and minor grooves in A- and B-helices it is obvious that a bulky spin label (as seen in the X-ray structure of TPA)13 is less well accommodated in the narrow major groove of an A-helix than in a B-helical form with a wide open major groove (helices modeled with PyMol, see ESI†).
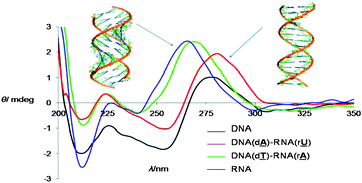 | ||
| Fig. 3 CD-spectra of spin labelled duplexes: left, A-; right, B-helix. | ||
Following this observation we argue that in the hybrid DNA(d![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) )–RNA(r
)–RNA(r![[U with combining low line]](https://www.rsc.org/images/entities/b_char_0055_0332.gif) ) the RNA seems to prefer more the B-like geometry, whereas the hybrid DNA(d
) the RNA seems to prefer more the B-like geometry, whereas the hybrid DNA(d![[T with combining low line]](https://www.rsc.org/images/entities/b_char_0054_0332.gif) )–RNA(r
)–RNA(r![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) ) remains in the RNA-like structure. As a further proof for our assignment we checked how RNase H1 recognizes these hybrids. RNase H1 lacks a consensus sequence for cleavage and preferentially cleaves 7–10 nucleotides from the 5′-RNA/3′-DNA terminus.14 It is known that preferred cleavage by the enzyme is a product of fitting sequential/helical geometry, predominantly A-like.15 We incubated the 6 helices, the DNA and RNA double strands as well as the four mixed hybrids and measured the cleavage rates by HPLC-analysis. For treatment with the enzyme RNase H (Bacillus) the double helices were formed in sterile water first. RNase H-buffer (50 mM Tris-HCl, 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol, pH 8.3) was added and the probes were incubated at 37 °C. The cleavage rate was followed by anionic exchange-HPLC (1 M LiCl).
) remains in the RNA-like structure. As a further proof for our assignment we checked how RNase H1 recognizes these hybrids. RNase H1 lacks a consensus sequence for cleavage and preferentially cleaves 7–10 nucleotides from the 5′-RNA/3′-DNA terminus.14 It is known that preferred cleavage by the enzyme is a product of fitting sequential/helical geometry, predominantly A-like.15 We incubated the 6 helices, the DNA and RNA double strands as well as the four mixed hybrids and measured the cleavage rates by HPLC-analysis. For treatment with the enzyme RNase H (Bacillus) the double helices were formed in sterile water first. RNase H-buffer (50 mM Tris-HCl, 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol, pH 8.3) was added and the probes were incubated at 37 °C. The cleavage rate was followed by anionic exchange-HPLC (1 M LiCl).
Neither DNA nor RNA was cleaved but both hybrids roughly at the same rate (see Table 1). In contrast the two spin labelled hybrid structures were cleaved at quite different rates to each other. The t1/2 for RNA(r![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) )/DNA(d
)/DNA(d![[T with combining low line]](https://www.rsc.org/images/entities/b_char_0054_0332.gif) ) is 26 min and for RNA(r
) is 26 min and for RNA(r![[U with combining low line]](https://www.rsc.org/images/entities/b_char_0055_0332.gif) )/DNA(d
)/DNA(d![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) ) is 148 min, a rate difference by a factor of approx. 6. Compared to the unmodified hybrids DNA/RNA (t1/2 120 min), the first combination is about 4.5× faster and the second 1.2× slower. We interpret this rate difference as being due to the fact that RNA(r
) is 148 min, a rate difference by a factor of approx. 6. Compared to the unmodified hybrids DNA/RNA (t1/2 120 min), the first combination is about 4.5× faster and the second 1.2× slower. We interpret this rate difference as being due to the fact that RNA(r![[A with combining low line]](https://www.rsc.org/images/entities/b_char_0041_0332.gif) ), where the spin label points to the minor groove, has no problem to adopt the A-helix geometry and is therefore cleaved nicely. In contrast for RNA(r
), where the spin label points to the minor groove, has no problem to adopt the A-helix geometry and is therefore cleaved nicely. In contrast for RNA(r![[U with combining low line]](https://www.rsc.org/images/entities/b_char_0055_0332.gif) ) there is limited space for the spin label in the major groove of an A form, thus it is more difficult to adopt the necessary A-helix geometry. This is in full agreement with crystal data for the RNase H1 in complex with RNA/DNA, where one finds a more flexible DNA and a more rigid RNA in the A-form.16 Furthermore for the DNA part it is known that acetylenic side chains in the 5-position of U enhance RNase H1 cleavage.17 Thus the RNA/DNA hybrid is more easily cleaved when the RNA is predominantly A-form and the DNA can be flexible enough to adopt the A-form as observed in the X-ray structure of the RNase H.16 This has consequences for the design of hybrid structures as in the case of antisense oligonucleotides, where strong A-form supporting nucleoside analogues inhibit RNase H cleavage.15 Similarly this may also be applicable to RNase H like enzymes as the argonaute silencing complex Ago2 (SLICER) in the RNAi pathway.18,19
) there is limited space for the spin label in the major groove of an A form, thus it is more difficult to adopt the necessary A-helix geometry. This is in full agreement with crystal data for the RNase H1 in complex with RNA/DNA, where one finds a more flexible DNA and a more rigid RNA in the A-form.16 Furthermore for the DNA part it is known that acetylenic side chains in the 5-position of U enhance RNase H1 cleavage.17 Thus the RNA/DNA hybrid is more easily cleaved when the RNA is predominantly A-form and the DNA can be flexible enough to adopt the A-form as observed in the X-ray structure of the RNase H.16 This has consequences for the design of hybrid structures as in the case of antisense oligonucleotides, where strong A-form supporting nucleoside analogues inhibit RNase H cleavage.15 Similarly this may also be applicable to RNase H like enzymes as the argonaute silencing complex Ago2 (SLICER) in the RNAi pathway.18,19
Conclusions
In summary we have introduced PELDOR in combination with CD for the characterization of RNA/DNA hybrid structures. We were able to show excellent agreement of PELDOR data with CD spectra. RNase H1 cleavage data support the observed structural differences. In agreement with the biological functionality of RNase H1 for faster cleaved hybrids an A-type structure should be present. This combination of PELDOR and CD shows promise for more detailed RNA/DNA structure determination.Notes and references
- (a) A. Rich, J. Biol. Chem., 2006, 281, 7693–7696 CrossRef CAS; A. Rich, Proc. Natl. Acad. Sci. U. S. A., 1960, 46, 1044–1053 CrossRef CAS; (b) J. T. Olimpo and J. J. DeStefano, Nucleic Acids Res., 2010, 38, 4426–4435 CrossRef CAS.
- H. Wu, W. F. Lima, H. Zang, A. Fan, H. Sun and S. T. Crooke, J. Biol. Chem., 2004, 279, 17181–17189 CrossRef CAS.
- J. Kurreck, Eur. J. Biochem., 2003, 270, 1628–1644 CAS.
- (a) A. D. Milov, A. B. Ponomarev and Y. D. Tsvetkov, Chem. Phys. Lett., 1984, 110, 67–72 CrossRef CAS; Y. D. Tsvetkov, A. D. Milov and A. G. Maryasov, Usp. Khim., 2008, 77, 515–550; (b) O. Schiemann, Methods Enzymol., 2009, 469, 329–351 CAS , Part B.
- (a) O. Schiemann and T. Prisner, Q. Rev. Biophys., 2007, 40, 1–53 CrossRef CAS; N. K. Kim, M. K. Bowman and V. J. DeRose, J. Am. Chem. Soc., 2010, 132, 8882–8884 CrossRef CAS; (b) A. Marko, D. Margraf, P. Cekan, S. T. Sigurdsson, O. Schiemann and F. T. Prisner, Phys. Rev., 2010, E81, 021911 Search PubMed; N. A. Kuznetsov, A. D. Milov, V. V. Koval, R. I. Samoilova, Y. A. Grishin, D. G. Knorre, Y. D. Tsvetkov, O. S. Fedorova and S. A. Dzuba, Phys. Chem. Chem. Phys., 2009, 11, 6826–6832 RSC.
- O. Schiemann, N. Piton, Y. Mu, G. Stock, J. W. Engels and T. F. Prisner, J. Am. Chem. Soc., 2004, 126, 5722–5729 CrossRef CAS.
- N. Piton, Y. Mu, G. Stock, O. Schiemann and J. W. Engels, Nucleic Acids Res., 2007, 35, 3128–3143 CrossRef CAS.
- G. Sicoli, G. Mathis, O. Delalande, Y. Boulard, D. Gasparutto and S. Gambarelli, Angew. Chem., Int. Ed., 2008, 47, 735–737 CrossRef CAS.
- (a) T. Strube, O. Schiemann, F. McMillan, T. Prisner and J. W. Engels, Nucleosides, Nucleotides Nucleic Acids, 2001, 20(4), 1271–1274 CrossRef CAS; (b) O. Schiemann, N. Piton, J. Plackmeyer, B. Bode, T. Prisner and J. W. Engels, Nat. Protoc., 2007, 2, 904–923 Search PubMed.
- S. A. Scaringe, F. E. Wincott and M. H. Caruthers, J. Am. Chem. Soc., 1998, 120(45), 11820–11821 CrossRef CAS.
- R. E. Martin, M. Pannier, F. Diederich, V. Gramlich, M. Hubrich and H. W. Spiess, Angew. Chem., Int. Ed., 1998, 37, 2834–2837 CAS.
- G. Jeschke, V. Chechik, P. Ionita, A. Godt, H. Zimmermann, J. Banham, C. R. Timmel, D. Hilger and H. Jung, Appl. Magn. Reson., 2006, 30, 473–498 CrossRef CAS.
- O. Frolow, J. W. Bats and J. W. Engels, Acta Crystallogr., Sect. E: Struct. Rep. Online, 2009, 65, o1848 CrossRef.
- P. S. Pallan and M. Egli, Cell Cycle, 2008, 7, 2562–2569 CrossRef CAS.
- W. F. Lima, J. B. Rose, J. G. Nichols, H. Wu, M. T. Migawa, T. K. Wyrzykiewicz, A. M. Siwkowski and S. T. Crooke, Mol. Pharmacol., 2007, 71, 83–91 CAS; W. F. Lima, J. B. Rose, J. G. Nichols, H. Wu, M. T. Migawa, T. K. Wyrzykiewicz, G. Vasquez, E. E. Swayze and S. T. Crooke, Mol. Pharmacol., 2007, 71, 73–82 CAS.
- M. Nowotny, S. A. Gaidamakov, R. J. Crouch and W. Yang, Cell (Cambridge, Mass.), 2005, 121, 1005–1016 CrossRef CAS; M. Nowotny, S. A. Gaidamakov, R. Ghirlando, S. M. Cerritelli, R. J. Crouch and W. Yang, Mol. Cell, 2007, 28, 264–276 CrossRef CAS.
- A. J. Gutierrez, M. D. Matteucci, D. Grant, S. Matsumura, R. W. Wagner and B. C. Froehler, Biochemistry, 1997, 36, 743–748 CrossRef CAS.
- Y. Wang, G. Sheng, S. Juranek, T. Tuschl and D. J. Patel, Nature, 2008, 456, 209–213 CrossRef CAS.
- W. F. Lima, H. Wu, J. G. Nichols, H. Sun, H. M. Murray and S. T. Crooke, J. Biol. Chem., 2009, 284, 26017–26028 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Details of the experimental procedures, UV, CD spectra, modeling and enzyme kinetics. See DOI: 10.1039/c0mb00258e |
| This journal is © The Royal Society of Chemistry 2011 |
