 Open Access Article
Open Access ArticleDesign, engineering and utility of biotic games†‡§
Ingmar H.
Riedel-Kruse¶
*,
Alice M.
Chung
,
Burak
Dura
,
Andrea L.
Hamilton
and
Byung C.
Lee
Department of Bioengineering, Stanford University, 299 W. Campus Drive, Fairchild, Room D243, Stanford, California 94305, USA. E-mail: ingmar@stanford.edu; Fax: +1 (650) 723 8544; Tel: +1 (650) 723 2380
First published on 18th November 2010
Abstract
Games are a significant and defining part of human culture, and their utility beyond pure entertainment has been demonstrated with so-called ‘serious games’. Biotechnology – despite its recent advancements – has had no impact on gaming yet. Here we propose the concept of ‘biotic games’, i.e., games that operate on biological processes. Utilizing a variety of biological processes we designed and tested a collection of games: ‘Enlightenment’, ‘Ciliaball’, ‘PAC-mecium’, ‘Microbash’, ‘Biotic Pinball’, ‘POND PONG’, ‘PolymerRace’, and ‘The Prisoner's Smellemma’. We found that biotic games exhibit unique features compared to existing game modalities, such as utilizing biological noise, providing a real-life experience rather than virtual reality, and integrating the chemical senses into play. Analogous to video games, biotic games could have significant conceptual and cost-reducing effects on biotechnology and eventually healthcare; enable volunteers to participate in crowd-sourcing to support medical research; and educate society at large to support personal medical decisions and the public discourse on bio-related issues.
Introduction
Games and play are deeply rooted in animal and human culture.1–3 Definitions of the term ‘game’ vary,4,5 but are generally associated with challenging human activities that have rules, goals, and uncertain outcomes. Often distinctions are made between simulations, toys, and puzzles5 – the following conclusions will generally apply to those as well. A key aspect of games is their entertaining, even addictive nature.5 Therefore, so-called ‘serious games’6,7 can provide effective means to motivate other objectives, such as sports games maintaining fitness or dice games teaching counting.Technological advancement constantly facilitates novel game modalities8 including their ‘serious’ applications. For example, computer technology has enabled video games9,10 that are now effectively utilized in physical therapy11 and higher-level education.12 Surprisingly, the current biotechnological revolution13 has had virtually no impact on gaming yet – let alone having generated any serious games. Therefore, we propose to actively investigate whether games based on biotechnology are feasible, what their distinct features are compared to existing game modalities, and what utility they could have for society.
As a conceptual framework, we suggest the term ‘biotic games’ ( = ‘of or relating to life, caused or produced by living beings’) as any activity including the necessary equipment (i) that falls under the concept of games,5 (ii) that has one or more humans interacting as active players with biological materials or processes, and (iii) where the game design and human experience depends on modern biotechnology. This framework excludes intentionally video games emulating biological processes electronically9 and evolutionary games,14 but acknowledges border-line cases such as games utilizing organic but non-biotic molecules, or games based on ‘ancient biotechnology’ like horse-polo.3 In short, we are interested in novel game experiences and applications that can be achieved with modern biotechnology.
In the following, we explore this concept in three classes of biotic games that we have designed. Our intention is to be thought provoking about how very different biological processes can be utilized and what novel game mechanisms can emerge. We characterize these games primarily under the following aspects: (i) The generalized rules defining the game: both man-made rules and implicit biophysical constraints; (ii) how the emergent features of these rules influence winning and play strategies; (iii) what unique biotic features differentiate them from existing games; and (iv) potential ‘serious’ applications. We conclude with a discussion on the broader technological and cultural implications of biotic games with an emphasis on their educational potential.
1st: Action games at the single cell level
Our first game is inspired by early action video games15 where players steer virtual objects on a screen (Fig. 1a). In our biotic game adaptation these objects are not virtual, instead they are living paramecia (Fig. 1b) contained inside a biotic game console (Fig. 1c). The human player interacts with these paramecia via a traditional game controller (Fig. 1d) and observes their responses on a video screen with a superimposed virtual game environment (Fig. 1e,h–l).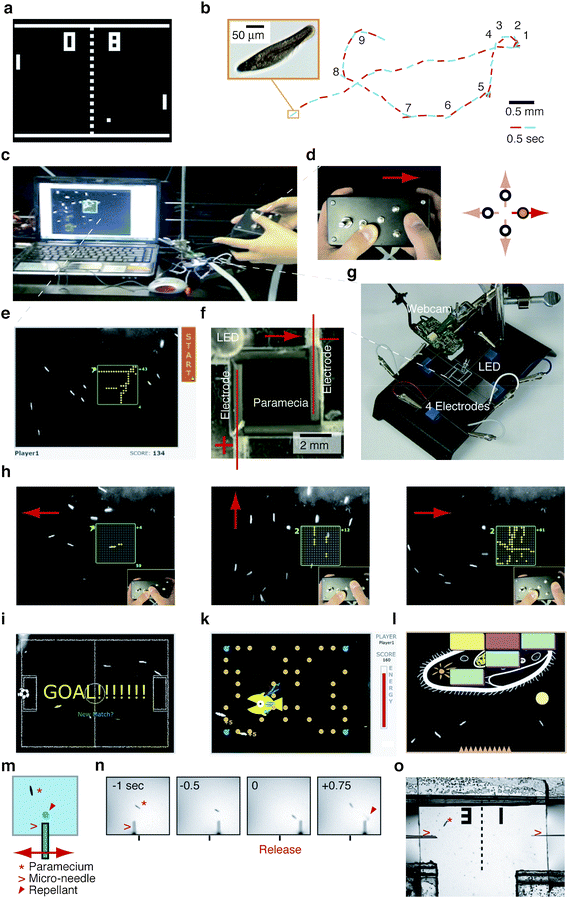 | ||
| Fig. 1 Biotic action games that are inspired by classic video games can be realized by controlling single-celled paramecia via electric fields or chemicals (see also ESI movies 1 and 2†): (a) Example of classic video game (PONG) where human players control virtual objects on a screen. (b) Paramecium (left inset) and its typical run-and-tumble track with frequent changes in swimming direction (#1–9). (c–g) Biotic game console: (c) overview; (d) hand held game controller for steering paramecia; (e) live image of paramecia streamed to computer screen with virtual game environment superimposed; (f) fluid chamber containing paramecia; (g) ‘biotic processor’ consisting of webcam, electrodes, connection to game controller, and fluid chamber. (h) Example sequence of game ‘Enlightenment’ demonstrating paramecia responses to the human player's actions. (i–l) Screenshots of other games using same set-up: (i) ‘Ciliaball’, where paramecia kick a ball into one of two goals. (k) ‘PAC-mecium’, where paramecia forage for yeast but have to escape a hunting zebrafish larvae. (l) ‘Microbash’ where paramecia destroy bricks to free the schematic drawing of a paramecium. (m) Alternate game mechanism where human player can influence paramecia by releasing chemicals from a micro-needle; player controls vertical position of needle also. (n) Example sequence of the game ‘Biotic Pinball’ (inspired by pinball) showing the successfully induced reversal of paramecium swimming direction after re-positioning the micro-needle and releasing a chemical repellant. (o) Two player version ‘POND PONG’ inspired by the classic video game ‘PONG’ (see a), where paramecia enter from below and two players aim to repel the paramecia with chemicals released from their respective needles towards the others player's side. | ||
Paramecia are ciliated, single-celled organisms that swim around in a run-and-tumble like motion by stochastically switching their swimming direction16–18 (Fig. 1b). Paramecia respond to external electrical fields by swimming towards the cathode, a phenomenon termed galvanotaxis19 (Fig. 1f, h). For the game setup, these paramecia are contained in a square fluid chamber, which has electrodes arranged along each side20 (Fig. 1f). The human player controls a swarm of these paramecia by applying electric fields along two axes via a hand-held device reminiscent of a conventional video game controller (Fig. 1d). The motion of these paramecia is captured with a webcam and displayed live on a computer screen (Fig. 1e, g, h). This set-up is turned into a game by overlaying virtual graphic objects onto the live video and defining how these virtual objects behave relative to the displayed paramecia. A game score is computed based on these interactions, which are ultimately influenced by the human player's actions (Fig. 1e, h).
This set-up enables the design of many different games of which we illustrate a few (see also ESI movie 1): In ‘Enlightenment’ (Fig. 1e, h)21 squares filled with ‘off-dots’ pop up on random sections on the screen. The human player scores points by steering paramecia through those dots and lighting them. In ‘Ciliaball’ (Fig. 1i) the paramecia kick a virtual soccer ball into one of two goals. In ‘PAC-mecium’ (inspired by ‘PAC-Man’)15 (Fig. 1k) the paramecia collect virtual yeast food and are occasionally bitten by a virtual zebra-fish larva. In ‘Microbash’ (inspired by ‘Breakout’)15 (Fig. 1l) the paramecia bounce a ball to remove virtual blocks, which eventually reveals the detailed schematic of a paramecium. These games have been test played by different people including visitors at a public event outside the lab. Adding fresh paramecia to the set-up takes about 5 min, and paramecia are typically playable for more than an hour. The chip is reusable after cleaning.
In these games, the novelty in play experience compared to traditional video games arises from the challenge of guiding multiple objects simultaneously and from the inherent biological nature of the system such as paramecia stochastically changing their swimming direction and often not responding to the applied stimuli as anticipated. Furthermore, the player's knowledge that these processes are real and not simulated influences players perception. For example, the question what these paramecia ‘feel’ while being steered was raised independently by different players.
Paramecia also respond to chemicals, a phenomenon termed chemotaxis,22 which motivated us to explore the integration of chemicals into these games as well. We constructed a set-up that enables the player to position and release chemicals from a micro-needle using a micro-manipulator. Analogous to the marbles in a pinball game, as paramecia swim towards the player they are ‘kicked back’, but instead of a mechanical impulse a physiological response induced by the repelling chemical drives this change in direction (ESI movie 2). Low Reynolds number hydrodynamics enable long-range non-contact interactions between paramecia and a needle.23 The relatively slow diffusion of the chemical18 allows a dynamic ‘repellant landscape’ to build up during the game. A two-player version inspired by the classic and first commercially successful video game PONG10 (Fig. 1o, ESI movie 2†) can be realized using two opposing micro-needles with the paramecium swimming in between. In their present form, these chemical games should be regarded as proof of concept as repeated play would require the removal of the released chemicals, which is left for more sophisticated micro-fluidic chip designs in the future. We call these games ‘Biotic Pinball’ and ‘POND PONG’, respectively.
We expect that such paramecia games (Fig. 1) would lend themselves well to microbiology and biophysics teaching given that paramecia are already common study material in American high-schools in a more observatory setting.24 As playing pinball conveys a sense of physical concepts such as gravity, inertia or spin, these paramecia games inform on micro-organismal behaviors, the biophysics of random-walks, diffusion, and low Reynolds number hydrodynamics.18 Such games are extendable to other microorganisms and alternate control mechanisms, such as photo-activate-able chemicals25 or direct light26 in more elaborate micro-fluidic environments and mazes. Students might be motivated to discuss and understand the observed phenomena in order to identify other winning strategies in such games,27 and multi-player games may increase competition and learning motivation.
2nd: Betting and logic games at the molecular level
Our second biotic game is inspired by horse racing28 (Fig. 2a) where players make educated bets on the order that specific horse-jockey pairings (C and X) reach a finish line. In the biotic game adaptation, players bet on when DNA amplified by specific PCR primer pairings (Fig. 2b) will reach a preset detection threshold during PCR (Fig. 2c).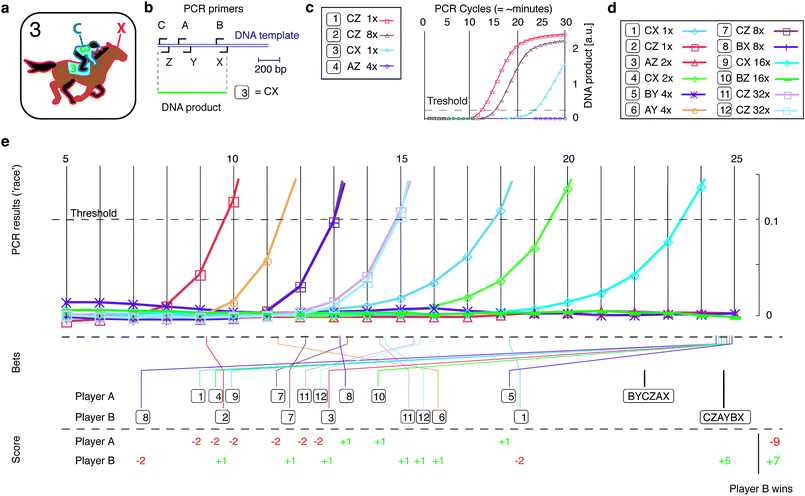 | ||
| Fig. 2 The molecular logic game ‘PolymerRace’ is inspired by horse race betting and has the goal to predict the speed of DNA amplification given specific PCR (polymerase chain reaction) primer pairings: (a) In a conventional horse race a jockey (C) and a horse (X) are paired as team #3, and spectators can place bets on their performance during a race. (b) Schematic for the binding positions of forward and reverse PCR primers (A, B, C, and X, Y, Z, respectively) on a DNA template. Selection of a primer pair (such as C and X, i.e., RXN 3), leads to a specific DNA amplification product during PCR. (c) Amount of amplification product increases with each PCR cycle and can be monitored in real time. Time to reach threshold depends on initial DNA template concentration (RXNs 1 vs. 2; 8× indicates a 8 fold dilution relative to maximal amount), and efficiency of primer combination (RXNs 1 vs. 3). If forward and reverse primer are in the wrong order (RXN 4), no DNA is amplified. Relative arrangement of primers as in (b). (d) Pseudo-random set of 12 primer pairs generated from set in b with systematically lower starting template concentration. (e) Example game played using set from d. Two players continuously monitor PCR results (upper panel), i.e. the ‘race’, and place bets in real time (middle panel). Bets specify cycle that chosen reaction will reach or has reached threshold (for example player B on RXNs 7 and 6, respectively). Players also specify the order of the six primers along the DNA template, i.e., BYCZAX and CZAYBX for player A and B (compare to b). Bets are scored after the race (lower panel) and player B wins. | ||
PCR (polymerase chain reaction) is a widespread molecular technique that uses a cyclic heating and cooling process to amplify DNA sequences specified by the binding locations of forward and reverse PCR primers on a DNA template16 (Fig. 2b). Individual reactions take place simultaneously inside a PCR thermo-cycler machine.16 The relative amount of DNA present in each reaction can be tracked in real time via a fluorescent signal (Fig. 2c). In a perfect reaction, this amount increases exponentially, doubling every cycle; thus an eight fold lower starting DNA concentration should reach threshold three cycles later (Fig. 2c; RXN 1 vs. 2; RXN = reaction). In practice, the number of cycles required to reach a given threshold not only depends on the initial DNA template concentration but also on reaction efficiency, which is influenced by reaction conditions and the length of the amplified DNA sequence (Fig. 2c, RXN 1 vs. 3). Additionally, if forward and reverse primers are incorrectly oriented, no template is amplified (Fig. 2b, c; RXN 4).
Our game is played with three forward and three reverse primers (Fig. 2b), from which a set of 12 reactions is prepared (Fig. 2d). Each reaction contains a random primer combination as well as a DNA template at progressively two-fold lower concentrations. Based on these conditions, individual reactions will reach threshold at different cycle numbers during the race; and some reactions will never reach threshold at all. Additionally, the PCR cycle conditions are set such that some products are not amplified with optimal efficiency.
At the game start, the two players are given the primer pairings, the DNA template concentrations, and which primers are forward and which are reverse, i.e., the full information in Fig. 2d. The order of primers relative to each other (Fig. 2b) is not revealed. The game starts and ends at cycles 5 and 24, respectively, which is approx. 20 min – a time that is naturally determined by the biochemical reaction conditions. PCR results are updated every cycle, (Fig. 2e, upper panel). Players use this real-time information to bet if and when a chosen reaction will reach threshold (Fig. 2e, middle panel). A bet consists of a player making public their prediction of at what cycle a specific reaction will reach threshold or if that reaction will not reach threshold by cycle 24. Each player can place only one bet for each reaction, and the opposing player cannot make the same bet but may bet on the same reaction reaching the threshold at a different cycle. Bets may be placed at any time during the game, including after a reaction has reached threshold (such as player B betting on RXN 11; Fig. 2e), and bets may not be retracted or changed. After the game, correct bets are awarded one point, and wrong bets are penalized two points (Fig. 2e, lower panel). The first player to correctly predict the relative order of all primers receives 5 additional points. The player with the most points wins. We call this game ‘PolymerRace’.
The play challenge (and fun) emerges from competition among players to be the first to place a given bet. With every cycle the players obtain new information about the relative order of the primers and the reaction efficiencies of individual primer pairings, all of which guide them in making predictions about the reactions. For example, if reaction efficiency was perfect RXN 7 should come up 3 cycles after RXN 2; in contrast RXN 1 is rather inefficient which has consequences for RXN 4. The results of RXNs 1–8 contain the full information to deduce the relative order of all primers in Fig. 2b (the proof is left to the reader). Thus, strategy emerges from the trade off between risky bets based on limited information and the danger of losing secure bets to aggressive or logic-driven players.
During the initial cycles of the game, players familiarize themselves with the reaction conditions (Fig. 2d). The following cycles are filled with processing information, placing bets and analyzing opponents' bets, which keeps players engaged throughout the entire game. Additionally, the PCR machine used here computes baseline corrections while running leading to shifts in displayed data-points (ESI movie 2†) and partially influencing the outcome of already placed bets. Combined with the error sources of the experiment (compare RXNs 11 and 12) this adds an element of chance and unpredictability to the game. Setting up a new game, i.e., preparing all reactions, takes about one hour, and the reagents and equipment involved lead currently to a non-negligible cost. Playing the game by watching a pre-recorded PCR-run (ESI movie 2†) alleviates these inconveniences.
Molecular games like ‘PolymerRace’ are suitable for undergraduate biology teaching as well as effective demonstration and training tools for PCR machines and other commercial biotechnological equipment – similar to the video game ‘Space War’ that was delivered along with the PDP1-computer in the 1960's.15 The pseudo-random reaction set used here (Fig. 2d) generates results (Fig. 2e) that enable players to measure reaction efficiencies based on dilution series (RXNs 2, 7, 11, and 1, 4, 9 for efficient and inefficient primer pairs, respectively) and experimental reproducibility of the used PCR machine (RXNs 11, 12). Therefore, such games can make an entertaining but also educational use of the lengthy run time by motivating the close observation and interpretation of what happens during the experiment.
3rd: Olfaction-strategy game at the multi-cellular level
Our third game adapts the iterated ‘Prisoner's Dilemma’.29 In this game, two players aim to maximize their own score according to a pay-off matrix by cooperating or defecting on the other player over repeated rounds (Fig. 3a). We utilize yeast (Fig. 3b) with their characteristic bread-vinegar-like smell to add an olfactory challenge to this game.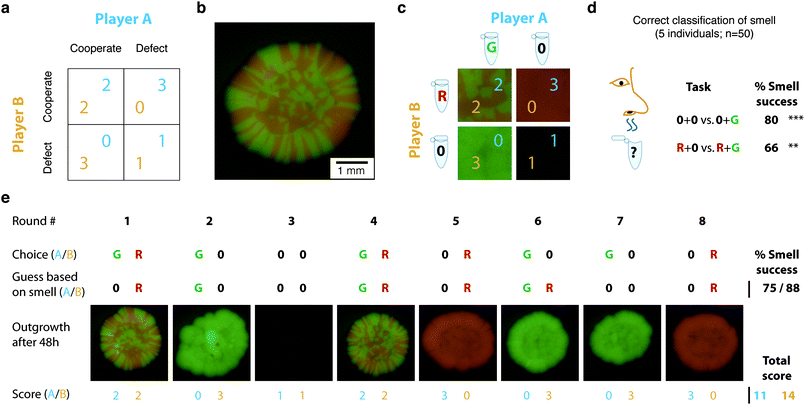 | ||
| Fig. 3 The multi-cellular game ‘The Prisoner's Smellemma’ utilizes colored yeast to integrate the olfactory senses into play: (a) Pay-off matrix of Prisoner's Dilemma. (b) Yeast colony grown from a mixture of two yeast strains expressing red (RFP) or green (GFP) fluorescent protein, respectively. (c) Game setup illustrating the two tubes each player has: One tube containing one of the colored yeast strains diluted into buffer, the other tube with buffer only; pay-off matrix as determined after mixing samples from each player and outgrowth over 48 h (compare to a). (d) Olfactory classification success for players on whether opponent has cooperated or defected depending on players own actions, i.e., (0 vs. 0 + G) or (R + 0 vs. R + G) if player defected or cooperated himself, respectively. (e) Example game played over eight rounds displaying the choices each player made, the guesses about the opponents actions based on the perceived smell, the results after colony outgrowth, and eventually the score. | ||
Before the game, each player receives one of two yeast strains diluted in buffer as well as a sample of buffer only (Fig. 3c). These yeast strains are genetically modified to express either a red or green fluorescent protein.16 This expression can be visualized by seeding a sample on an agar plate and incubating it for 1–3 days. When colonies are mixed, a red–green patterned yeast colony emerges (Fig. 3b) due to a competitive growth process.30 During each round of the game, both players choose to contribute equal volumes of either their yeast strain (‘cooperating’) or the pure buffer (‘defecting’) to a common empty tube with their score depending on a pay-off matrix (Fig. 3c). Since the game is played over multiple rounds, players need to infer whether the opponent is cooperating or not. In contrast to the conventional version of the prisoner's dilemma this information is not revealed directly to the other player, instead both players have to gather that information purely by smelling on the well-mixed tube.
We found that humans can smell the difference between opponent adding yeast or pure buffer (Fig. 3d). The success rate is not perfect, but significantly above chance thereby providing a play challenge. The trend that correlates classification success with relative concentration change may be expected from human olfactory psychophysics experiments.31 Determining the opponent's choice based on smell represents noisy information29 and guides players in their strategy for subsequent rounds (Fig. 3e, upper part). After the game, the result of the player's choices (Fig. 3e, lower part) is revealed unambiguously by seeding a sample from each round's tube on an agar plate. The results of the various yeast colonies after outgrowth can then be matched to the pay-off matrix (Fig. 3c). A single game lasts eight rounds. The goal is to gain the highest score in a multi-player tournament rather than winning a single game. The game can be set-up within 5 min and takes about 10 min to play. We call this game ‘The Prisoner's Smellemma’.
There are less time-consuming ways to confirm the players choices than growing colonies, yet the resulting red–green patterns (Fig. 3b,e) might inspire the design of other games where the spatio-temporal growth processes would enable game dynamics related to Go, Reversi, or Hex.32 Engineering the required genetic networks would provide interesting applications for synthetic biology33 or new pattern formation paradigms for developmental biology and ‘evolutionary games’.14 (Note the significantly different usage of the term ‘game’ as humans may act as objective observers or actual players.5) ‘The Prisoner's Smellemma’ also demonstrates how biotic games facilitate the natural integration of the human chemical senses into play, while conventional games are almost exclusively restricted to the visual, acoustic and mechanical senses.5 Olfactory games have many applications, for example widening the joyful life experience of visually or acoustically impaired people, or enabling self-monitoring for neurological diseases like Alzheimer's or Parkinson, where severe olfactory dysfunctions have been reported to appear much earlier than those on cognition or movement.34
Discussion: Future potential and impact of biotic games
These three biotic games (Fig. 1–3) demonstrate play-ability from the molecular to the multi-cellular scale, and we expect future biotic games to provide even more distinct gaming mechanisms compared to existing modalities – the complexity and variety of today's video games were similarly unforeseeable at their beginnings.15On what time scale could we expect such games to mature and have a significant impact? More than five decades of video game history provide a base for extrapolation:9,10,15 One of the first electronic games, ‘Tic-Tac-Toe’, was implemented in 1952 on an early vacuum-tube based computer with the goal of demonstrating and studying human-computer interactions.9 Interestingly, the only bioengineered game we know of is also ‘Tic-Tac-Toe’, developed in 2003 as a paradigm for deoxyribozyme-based computation.35 Arguably the first true video game, thereby creating a novel game mechanism, was ‘Tennis-for-two’ on an oscilloscope in 1958,9 which entertained visitors of the Brookhaven National Laboratory, and which could be paralleled to the games in Fig. 1h–l. It took nearly two more decades until video games rose to culturally significant phenomena in the 1970's in the developed world via arcade machines, video consoles, and microcomputers.9 For biotic games this should happen significantly faster since their underlying technologies seem to advance at a higher rate such as the integration density on micro-fluidic devices beats Moore's law for electronic circuits,36 since conventional computer and web technology nowadays could take over non-essential ‘biotic features’ of games (Fig. 1i–l), and if their development were pursued more actively than was done initially for video games9,15
As with video games, biotic games could become significant economic, technological, and scientific drivers:9,10,15 Biotic games could enforce technological price drops that positively feed-back onto healthcare and research investments37 – imagine PCR machines as common household applications for gaming and recognizing spoiled food – similarly unthinkable as personal computers 50 years ago and which were often purchased for gaming purposes.9 Technologies primarily developed for gaming could eventually find more ‘serious’ applications – similarly as graphics processing units (GPU) developed for real-time video game effects now enable molecular dynamic simulations.38 Volunteers could also be attracted into ‘biotic crowdsourcing’ to aid tasks like combinatorial drug screening39 not only by performing data analysis online40,41 but by actually running experiments remotely – similar to solving protein structures or labeling images with the electronic online games ‘FoldIt’42 and ‘ESP’,43 respectively.
Biotic games have multiple stand-out features compared to existing game modalities: The integration of biological materials and processes below the milimeter scale, the inclusion of olfactory senses, the usage of biological variability and noise44 as a natural ‘dice’, and the provision of a true reality experience rather then a virtual one.45 Immersing video games,27 which show increasing success in virtualizing biological worlds and experiences, necessarily fall short in a few aspects compared to biotic games: (i) The currently available computational power poses challenges to realistically emulate even modest biological phenomena, such as the swimming of ciliates at low Reynolds numbers.46 (ii) The details of biological systems are not completely understood, hence the realism is limited by the game designers knowledge. (iii) The experience of holding a pipette or interaction with real chemicals or biological systems still provides a different human experience. (iv) Certain applications like medical drug screening games (see above) inherently require the interaction with true biology. Since game play always explores the rules and laws of the world that the game is set in,5 biology teaching might be best set within a truthful biological world, with the gaming situation providing a natural motivation for exploration. Additionally, we expect that implementing the same game electronically and biologically will lead to cross-fertilization between both disciplines thereby helping to improve the creation of virtual worlds as well as understanding how biological systems behave and function.
We also envision an educational impact beyond the student level for society as a whole. The modern life sciences constantly lead to new diagnostic and treatment options that require informed decisions from patients,47 and modern biotechnology can raise substantial public controversies, such as on genetically modified foods, stem cell research, or teaching evolution.48–51 Dealing with these issues is challenging as many people have insufficient experience and knowledge about the underlying biological key concepts that are often abstract and unclear.52 This is in contrast to the integration of computer technology in the developed world. Even children interact with this technology in playful ways,10 and many of today's self-taught computer experts started out as dedicated video game players.10 Equivalently, biotic games would enable a significantly higher portion of the population to interact directly or online with biological processes at those small scales. This first hand experience could make them more informed, comfortable, and conscious about what modern biotechnology is, and what its true risks and benefits are. This is a necessity when it comes to personal medical choices or democratic policy making on controversial bio-related topics in order to meet global health, food, and environmental challenges.53,54
And finally, we hope that biotic games will be played for fun.
Methods
Paramecia culture
Paramecium caudatum was obtained from Biological Resource Center 8030 85th St, Amery, WI 54001 and ordered through Mountain Home Biological Inc., P.O. Box 277, White Salmon, WA 98672. Paramecia were either used directly within 5 days after delivery, or from a colony established in the lab.17 Different methods were used to concentrate Paramecia: For example, placing 50 ml of the paramecia stock solution into a Nunc vacuum filter (care was taken not to take too much dirt which typically had settled to the bottom) and about 90% of the buffer was sucked away (termed ‘standard buffer’). Alternatively, electrodes spaced 1 cm apart where placed into the paramecia stock solution and 6V were applied; after a few minutes paramecia concentrated near the cathode and were sucked up with a pipette. Paramecia were then kept at their concentration or later diluted back into standard buffer or Tris-buffer (1 mM Tris, 1 mM Citric acid, 1 mM CaCl2, pH = 7.017).Electrical control of paramecia
The fluid chamber (Fig. 1f) with dimensions 10 mm × 10 mm was made out of 1.5 mm thick acrylic using laser ablation. Elongated holes were rasterized at the four corners of the chamber using laser ablation to enable insertion of the four electrodes (Fig. 1f, g). The chamber was glued on microscope slides (WVR) using an Instant Adhesive (PartsMasterTM). Four carbon electrodes with 0.9 mm diameter (Pentral; typically used for pens) were inserted through the rasterized holes. Carbon electrodes are more bio-compatible than metal ones. The chamber was sealed off on all sides with epoxy to avoid leakage and stabilize electrode positions. Paramecia were loaded into the chamber with a transfer pipette. A cover glass was occasionally used to seal the chamber against evaporation. Paramecia were typically refreshed after 3–4 h.A PCB (printed circuit board) was prepared to serve as housing stage for the fluid chamber and to facilitate the electric connection between chamber and game controller. The PCB consisted of LED for illumination, eight crocodile clips for electrode connections and a 10-pin header connector for receiving the push button connections on the game controller (Fig. 1d). The layout of the PCB was drawn by hand using a Sharpie black marker on the copper side, and then the PCB was put into a Ferric Chloride solution (MG Chemicals) to etch away extra copper. The backside of the PCB board faced the fluid chamber and was painted with a Sharpie black marker to provide an anti-reflective background for dark field imaging. Two L-shaped restraints were glued on the backside of the PCB to fix the position of the chamber on the PCB stage. For game play, the chamber was placed in between the L-shaped restraints and electrodes were connected to the PCB board by crocodile clips.
The game controller consisted of four push buttons (Digikey, CKN1639-ND) to control the field direction along two axes (Fig. 1d), an on/off switch (Digikey, 450-1521-ND) to turn on the LED (Linrose, BCMD333UWC) on the PCB stage and a 2 K-ohm potentiometer (Fry's Electronics, NTE 502-0104) to arrange the intensity of the LED. The LED was connected to 6 V (3 V Enercell batteries in series) through the on–off switch, the potentiometer and a 100 ohm resistor. When pressed, push buttons connected the 3V batteries to generate fields of 3 V cm−1, which is about 3 times more than required to evoke responses,19 and 3 times lower than what would significantly reduce the lifetime of the paramecia.20 One pair of electrodes was always short-circuited when the other pair was activated, which minimized charge build-up at electrodes and led to more precise control of the paramecia. All electrical components were soldered on a prototype board (RadioShack, Model:276-148). The game pad hardware was enclosed in a black project box (RadioShack, Model: 270-1803) and push button connections on the game pad were transferred to the PCB stage (on which the chamber was placed) by a ribbon cable and an idc-socket.
The paramecia were imaged and recorded with a standard commercial webcam (Logitech C905). The webcam was first dissembled and an additional 5–10× lens (RadioShack) was put in front of the webcam lens. The webcam was mounted on a standard clamp stage and positioned for optimal focus (Fig. 1g). The light coming from the LED entered the chamber nearly horizontally, leading to dark-field illumination with bright paramecium on a black background (Fig. 1h). Live video was streamed to a PC laptop, and the imaged region and illumination condition were optimized via the camera control software that had been supplied with the webcam.
Flash game interface
Custom-software for the games was written using Adobe Flash CS5. The display of the game screen comprised of the live video monitoring the fluid chip and the superimposed virtual game graphic objects. At the beginning of each game, still images of the live video were captured for 6 s at 30 fps and averaged to produce a background image for motion tracking of paramecia. During game play images were acquired at 10 fps, from which the background image was subtracted. Rescaling and thresholding of the subtracted images reduced noise for detecting changed pixel value within the images, i.e., high pixel values corresponded to paramecia. Each virtual flash objects then assays whether it coincides with high pixel values, and if so, triggering corresponding actions, for example moving the virtual ball (Fig. 1i).Chemical control of paramecia
2,4-Dichlorophenoxyacetic acid (Sigma-Aldrich) (short: 2,4-D)17 was used as repellant at a stock concentration of 2 mM in ddH20. 2,4-D works as a repellent for Paramecia,17 which was qualitatively confirmed using a T-maze assay, as well as with the actual games (ESI movie 2†). To visualize the repellant it was mixed 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with 5% Phenolred (Sigma-Aldrich) in 5mM Hepes.
1 with 5% Phenolred (Sigma-Aldrich) in 5mM Hepes.
Micro-needles (World Precision Instruments; Item NO. 4878) were pulled on a micropipette puller (P 80/PC; Sutter Inst. Co.). Needles were broken open at the tip using a forceps with an approximate opening diameter of a typically injection needle (5 μm) – although significantly smaller ones worked also. Mineral oil was injected with a syringe from the backside (large opening) and the needle was mounted onto an oil-driven injector (Nanoject II Auto-Nanoliter Injector; Drummond Scientific). The injector was held by a 3-axis micromanipulator (Merzbacher). Then the needle was filled with the dyed 2,4-D solution.
Fluid chambers were made by placing micro cover glass (VWR) on Microscope Slides (Fischer) separated by four layers of double-sided Scotch tape. Cover glass and tape was cut to size with a diamond cutter and a razor blade, respectively. Different chamber designs were realized (ESI movie 2†). The solution with paramecia was loaded into one side of the fluid chamber, and the ‘standard buffer’ into the second one, with an air gap between the two. The needle with repellent was placed into this gap (ESI movie 2†). At the start of the game this gap was bridged with an additional drop of buffer. Individual games required significant preparation time as chambers were often not reusable. Additionally, games varied in play experience as timing and number of paramecia entering the playfield relied on their random behavior. Hence the presented results (ESI movie 2†) should be regarded as proof of principle with a game setup as reliable as the one using the electrical control awaiting future work.
Games were either played by direct observation through the binoculars or by watching the simultaneous video camera recording on a screen. Movies and images were taken on two different systems: An AxioCam HRc (Zeiss) with the acquisition software AxioVision (Zeiss) on an Olympus MVX10, and an AxioCam HRm (Zeiss) with the acquisition software AxioVision (Zeiss) on an AxioImager (Zeiss) – in both cases using bright field illumination. Movies were taken with different resolutions and frame rates of about 20 fps. Game scores (ESI movie 2†) were overlaid afterwards on the movie by hand.
PCR
A 5260 bp sized pCS2 plasmid with an H2B-cerulean insert was linearized at the Not1 cut site. Plasmid was used at a working concentration of 3.4 ng μl−1, and stored at −20 °C. Primers were designed using Genious 4.7.4 (Biomatters). Inside a continuous part of the pCS2 region successive blocks of 200 bp were assayed for the best possible primer pair that had a melting temperature between 57 and 58 °C, a primer size between 20 to 40 pb, a GC content between 40 and 60%, a GC clamp of 2, and a resulting product size above 70. Among those the following 6 primers were chosen (Fig. 2b): F_0036 - CTTTTGTTCCCTTTAGTGAGGGTTAATTGC; R_0140 - CGTATGTTGTGTGGAATTGTGAGCG; F_0249 - CTGCATTAATGAATCGGCCAACGC; R_0390 - CCGTATTACCGCCTTTGAGTGAGC; F_0692 - GTAGGTATCTCAGTTCGGTGTAGGTCG; R_0799 - ACTCAAGACGATAGTTACCGGATAAGGC. Nomenclature indicates directionality and distance in base-pairs relative to the cut site. Primers were ordered from IDT pre-diluted in 100 μM IDTE Buffer pH 8.0. Primers were further diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 in distilled water, aliquoted, and stored at −20 °C. All primer combinations were tested to give specific products of expected size under optimal conditions.
10 in distilled water, aliquoted, and stored at −20 °C. All primer combinations were tested to give specific products of expected size under optimal conditions.
PCR reactions were assembled on ice using the Rotor-Gene SYBR Green PCR kit (#294972, Quiagen). Each primer was at a final concentration of 1 μM. DNA was at progressively lower concentrations (Fig. 2d) with the highest concentration of 0.005 ng μl−1, and RNAase free water to reach a final volume of 25 μl. PCR was run on a Mastercycler ep realplex 4 (Eppendorf) with 5′ at 95 °C, followed by 30 two-step cycles of 10′ at 95 °C and 20′ at 60 °C, which is approx. 1 min per cycle. To minimize cost of reagents and setup-time a variety of games were carried out on a custom made MATLAB (Mathworks) program simulating the PCR machine. Furthermore, movies were taken from the display of the PCR machine during the run of a game (ESI movie 3†) and then played back to players who had not seen the data set before. The betting during the game was tracked with labeled 1.5 ml eppendorf-tubes (ESI movie 3†).
Yeast
Two Yeast strains (s. cerevisiae) ubiquitously expressing GFP or RFP, were grown and maintained in YPD media (5 g yeast extract, 10 g peptone, 10 g dextrose, in 500 ml water) at 28 °C and 4 °C, respectively. 1 h before game play, 1 ml of each Yeast strain was diluted in PBS in a 50 ml Eppendorf tube giving an optical density (OD) of 0.8 and kept at room temperature. Each player then received the corresponding strain, and another 50 ml tube with pure PBS. During each round of the game both players added 500 μl from one of their tubes to a 1.5 ml Eppendorf tube. The tube was colored to prevent visual inspection of the liquid. The tube was gently mixed and then both players were allowed to smell. Which player smelled first alternated between rounds. After all rounds of a single game, 0.5 μl from each tube was seeded on an YPD agar plate (Teknova). The plate was incubated for 24 h at 28 °C and imaged under red and green fluorescence (Leica M205 FA/DFC 500). If larger scale patterns for visual effect were desired (Fig. 3b), solutions were diluted up to 300× with PBS and grown correspondingly longer.Before the actual game, players were often familiarized with the iterated ‘Prisoner Dilemma’ by playing the standard version against each other on paper or against an artificial player found online. Furthermore, two sets of smell tests with 10 randomized tubes each were done that mimicked the smell test encountered during the game: One set had either nothing of half the yeast concentration, and the other half or the full concentration (compare smell test in Fig. 3d).
Acknowledgements
We would like to thank the Elowitz, Fraser, and Laurent labs at Caltech and the Clandinin and Dolmetsch labs at Stanford for generously sharing equipment and reagents. We are particularly grateful in various ways to Long Cai, Tom Clandinin, Rhiju Das, Franz Ferdinand, David Koos, Jeffrey Tsao, Henry Lowood, Alexey Pajitnov, Stephen Quake, Daniel Schwartz, and Adrien Trieulle as well as members of the Stanford Bioengineering department. This work was supported by the Beckman, Powell, and R & P Anderson foundations. We apologize to authors who were not cited for space constraints. We dedicate this work to Martin Gardner (1914–2010).References
- J. Goodall, In the Shadow of Man, Mariner Books, New York, 2000 Search PubMed.
- O. E. Wilson, Sociobiology: the new synthesis, Belknap Press of Harvard University Press, Cambridge, 2000 Search PubMed.
- S. Craig, Sports and Games of the Ancients, Greenwood, Santa Barbara, 2008 Search PubMed.
- C. Craford, The Art of Computer Game Design, Osborn/McGraw-Hill, New York, 1984 Search PubMed.
- K. Salen, E. Zimmerman, Rules of play: game design fundamentals, MIT Press, Cambridge, 2003 Search PubMed.
- C. Abt, Serious Games, University Press of America, Lanham, 1987 Search PubMed.
- J. Schollmeyer, Bulletin of the Atomic Scientists, 2006, 62, 34 Search PubMed.
- S. Scheri, The casino's most valuable chip: how technology transformed the gaming industry, Institute for the History of Technology, 2005 Search PubMed.
- M. J. P. Wolf, 2008, The Video Game Explosion: a History from PONG to Playstation and Beyond, Greenwood Press, Westport Search PubMed.
- M. Mertens, Wir waren Space Invaders: Geschichten vom Computerspielen, Eichborn Verlag, Frankfurt am Main, 2006 Search PubMed.
- B. Lange, S. M. Flynn and A. A. Rizzo, Eur J Phys Rehabil Med, 2009, 45, 143 Search PubMed.
- M. J. Mayo, Science, 2009, 323, 79 CrossRef CAS.
- S. Chien, Y. C. Fung, An Introductory Text to Bioengineering, World Scientific Publishing, Hackensack, 2008 Search PubMed.
- M. A. Nowak and K. Sigmund, Science, 2004, 303, 793 CrossRef CAS.
- S. L. Kent, The ultimate history of video games, Prima Publishing, Roseville, 2001 Search PubMed.
- B. Alberts, Molecular biology of the cell, Garland Science, 2008 Search PubMed.
- N. Takiguchi, et al. , J Biosci Bioeng, 2002, 93, 416 CAS.
- H. C. Berg, Random walks in biology, Princeton University Press, Princeton, 1993 Search PubMed.
- R. Eckert, Science, 1972, 176, 473 CrossRef CAS.
- K. Takahashi, N. Ogawa, H. Oku, K. Hashimoto, Proceedings of the 2006 IEEE International Conference on Robotics and Automation1408, 2006 Search PubMed.
- I. Kant, Berlinische Monatsschrift, 1784, 481 Search PubMed.
- J. Van Houten, Science, 1979, 204, 1100 CrossRef CAS.
- L. D. Landau, E. M. Lifshitz, Fluid Mechanics (Course of Theoretical Physics), Elsevier, Amsterdam, 1959 Search PubMed.
- L. M. Sepel, E. L. Loreto and J. B. Rocha, CBE Life Sci Educ, 2009, 8, 338 Search PubMed.
- S. Khan, et al. , Biophys. J., 1993, 65, 2368 CrossRef CAS.
- K. J. Hellingwerf, Antonie van Leeuwenhoek, 2002, 81, 51 CrossRef CAS.
- C. Dede, Science, 2009, 323, 66 CrossRef CAS.
- R. Eng, Betting on Horse Racing for Dummies, Wiley Publishing, Inc, Hoboken, Nj, 2005 Search PubMed.
- B. Brembs, Oikos76, p. 14 Search PubMed.
- O. Hallatschek, P. Hersen, S. Ramanathan and D. R. Nelson, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 19926 CrossRef CAS.
- A. Keller and L. B. Vosshall, Current Biology, 2004, 14, R875 CrossRef CAS.
- D. Sidney Parlett, The Oxford history of board games, Oxford University Press, Oxford, 1999 Search PubMed.
- S. Basu, Y. Gerchman, C. H. Collins, F. H. Arnold and R. Weiss, Nature, 2005, 434, 1130 CrossRef CAS.
- C. Hawkes, Adv. Otorhinolaryngol., 2006, 63, 133 Search PubMed.
- M. N. Stojanovic and D. Stefanovic, Nat. Biotechnol., 2003, 21, 1069 CrossRef CAS.
- R. M. Frederickson, Chem. Biol., 2002, 9, 1161 CrossRef CAS.
- H. Becker, Lab Chip, 2009, 9, 2119 RSC.
- J. A. Anderson, C. D. Lorenz and A. Travesset, J. Comput. Phys., 2008, 227, 5342 CrossRef.
- A. Peters, D. M. Brey and J. A. Burdick, Tissue Eng. Part B: Reviews, 2009, 15, 225 Search PubMed.
- S. Ekins, A. J. Williams, Pharm. Res., published online, 2010 Search PubMed.
- T. I. Oprea, et al. , Nat. Chem. Biol., 2009, 5, 441 CrossRef CAS.
- S. Cooper, et al. , Nature, 2010, 466, 756 CrossRef CAS.
- L. von Ahn, L. Dabbish, ACM Conference on Human Factors in Computing Systems, CHI319, 2004 Search PubMed.
- U. Alon, An Introduction to Systems Biology: Design Principles of Biological Circuits, Chapman & Hall/CRC, Boca Raton, 2006 Search PubMed.
- M. Sincell, Science, 1999, 286, 398 CrossRef CAS.
- G. J. P. Stephen, M. King, Ed., Analysis of the Ciliary/Flagellar Beating of Chlamydomonas (Elsevier Academic Press, San Diego, 2009) Search PubMed.
- R. Klitzman, Genet. Med., 2009, 11, 880 Search PubMed.
- Paul Bert, D. Baltimore, S. Brenner, R. O. Roblin and M. F. Singer, Proc. Natl. Acad. Sci. U. S. A., 1975, 72, 1981 CrossRef.
- M. W. Bauer, G. Gaskell, Biotechnology: The Making of a Global Controversy, Cambridge University Press, Cambridge, 2002 Search PubMed.
- J. G. Reich, Science, 2002, 296, 265 CrossRef CAS.
- H. I. Miller, P. Morandini and K. Ammann, Trends Biotechnol., 2008, 26, 122 CrossRef CAS.
- G. Gaskell, et al. , Nat. Biotechnol., 2000, 18, 935 CrossRef CAS.
- M. Berger, Public Health and Agricultural Biotechnology: A Review of the Legal, Ethical, and Scientific Controversies Presented by Genetically Altered Foods, Universal-Publishers, 2000 Search PubMed.
- H. C. J. Godfray, et al. , Science, 2010, 327, 812 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Movies 1 to 3. See DOI: 10.1039/c0lc00399a |
| ‡ Author contribution and order: IHRK conceived the project and wrote the paper. All authors contributed ideas and participated in game testing, with primary contributions: AMC developed flash code Fig.1h–l; BD and BCL developed setup Fig. 1c–g; ALH carried out experiments Fig. 2 and 3; IHRK contributed to all experiments, developed set-up Fig. 1m–o. First author highest contribution, other authors ordered alphabetically by last name. |
| § Conflict of interest: IHRK has filled a provisional patent application. |
| ¶ Previous address: Division of Biology, California Institute of Technology, Pasadena, California 91125, USA. |
| This journal is © The Royal Society of Chemistry 2011 |
