Materials chemistry in the emerging field of synthetic biology
Cameron
Alexander
*a and
Rachel K.
O'Reilly
b
aThe School of Pharmacy, Boots Science Building, University of Nottingham, University Park, Nottingham, NG7 2RD, UK. E-mail: cameron.alexander@nottingham.ac.uk; Fax: +44 (0) 115 951 5102; Tel: +44 (0) 846 7678
bDepartment of Chemistry, University of Warwick, Gibbet Hill Road, Coventry, CV4 7AL, UK
In general terms, synthetic biology can be split into two central paradigms. Each has specific features and it is worth mentioning two keynote papers to exemplify each method.
In one approach, termed the ‘top-down’ method by analogy with micromachining, pre-existing biological systems are re-engineered via modification or replacement of individual components or gene circuits, such that a new function is generated. The most well-known example of the ‘top-down’ method is from Gibson et al.,1 in which a mycobacterial ‘chassis’ was used to house an entirely synthetic genome. In this case the synthetic chromosome was specifically encoded with functional (β-galactosidase) and labeling (‘watermark’) sequences in order to enable facile discrimination from wild-type mycobacteria.
The second approach seeks to instill biological or biomimetic features from assembly of functional components, which may or may not be biological in origin. This is named the ‘bottom-up’ method, again in analogy with nanotechnology terminology. Perhaps the leading example of this was reported by Mansy and co-workers,2 who developed protocells from fatty acid ester vesicles. These were used to encapsulate nucleotide templating and copying components, but, importantly, were permeable to nucleotides, but impermeable to the templated polymers produced. In effect, these protocells behaved as obligate heterotrophs, i.e. they were able to metabolise exogenous factors while retaining their products.
These two papers indicate some very significant advances in the two main strands of synthetic biology, but there remain many scientific and technical challenges before generic synthetic cell constructs or protocells emerge or practical applications are feasible. These include better manipulation of biological circuits and complex gene constructs, enhanced control in preparation of synthetic containers, improved understanding of transport across natural and synthetic membranes, and new means for characterization of complex multi-component systems. The papers in this themed issue cover many of the above aspects of synthetic biology and point to real progress in the fundamental science. In addition, the review by Stano and co-workers (P. Stano et al., J. Mater. Chem., 2011, 21, DOI: 10.1039/c1jm12298c covers in greater detail the materials chemistry of compartmentalized reactions in lipid vesicle protocells.
The manipulation of complex functional biological components is addressed by Y.-H. Percival Zhang and colleagues (Fig. 1). The thematic area of this work is in cell-free synthetic pathway biotransformation (SyPaB), a methodology which might enable the production of complex materials from simple building blocks but in a highly controllable manner. Central to SyPaB is the concept of modifying natural ‘machinery’ (enzyme complexes, gene circuits) in such a way as to alter feedstock and product profiles to desired materials. Multiple enzyme cascades can be used for chemical transformations, but conditions must be optimised so that denaturation does not take place, or so that undesirable by-products do not build up and inhibit the intended reaction processes.
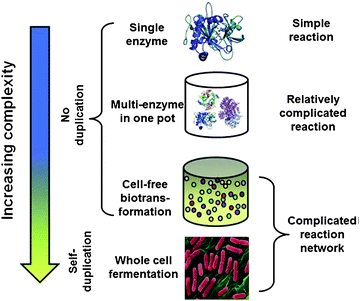 | ||
| Fig. 1 Comparison of biocatalysis mediated by a single enzyme, multiple enzymes, cell-free synthetic enzymatic pathway, and whole cells, in an order of increasing complexity. See Y.-H. P. Zhang et al., J. Mater. Chem., 2011, 21, DOI: 10.1039/c1jm12078f. | ||
Engineered enzymes may be needed for cell-free transformations, either for altered substrate/product profiles, or for enhanced stability in the acellular environment. In addition, immobilisation chemistries for enzymes used at heterogeneous surfaces or in supported hydrogels will need to be enhanced in order to ensure that the intended reaction sequences take place in the correct timeframe and yields. Zhang and co-workers describe progress in this area of synthetic biology, particularly related to enzyme co-factor use and regeneration and process design.
Protein materials chemistry is also a focus for Uhlenheuer et al. (D. A. Uhlenheuer et al., Strong supramolecular control over protein self-assembly using a polyamine decorated β-cyclodextrin as synthetic recognition element, J. Mater. Chem., 2011, 21, DOI: 10.1039/c1jm12736e). In this case, as an early stage example of designed biomolecular machinery, the modification of various fluorescent proteins is described. By means of modified cyclodextrin and lithocholic acid ‘handles’, the linkage of two different proteins (monomeric cyan-fluoresecent and monomeric yellow-fluorescent protein; mCFP and mYFP) is explored. Binding of the steroid group attached to mCFP within the hydrophobic cavity of cyclodextrin attached to mYFP takes place readily and FRET measurements enable this interaction to be studied in the presence of an inhibitor – in this case a cyclodextrin modified with side-chain cysteamines. Further cyclodextrin-derivatised proteins are then introduced to modify assembly and disassembly of mCFP and mYFP conjugates, enabling a FRET analysis of protein dimerization that maps out the stages of interaction. The significance of this work is that it establishes a facile means of monitoring the assembly of potential synthetic biology building blocks derived from materials other than nucleic acids. Many other complexing systems could also be used and are under active investigation,3 but the simplicity and modular nature of this approach offers much promise in ‘bottom-up’ assembly.
A further aspect of interest in this emerging field is in evaluating the modulation of essential biological events using synthetic materials. In this themed issue the paper ‘Influence of pegylation on peptide-mediated liposome fusion’ by Alexander Kros, DOI: 10.1039/c1jm11722j explores this through examining the effect on established liposome fusion mechanisms through liposome surface modifcation with complementary peptides and PEG (Fig. 2).
 | ||
| Fig. 2 Conceptual illustration of the proposed mechanism of fusion inhibition by pegylated lipids. Surface-bound PEG chains prevent the close contact of membranes by their steric effect, thus the peptide complexes are destabilized. Therefore, the population of docked liposomes is significantly reduced, inhibiting liposome fusion. | ||
In this report the authors highlight that a different mechanism than liposome fusion (through directed complex formation) is needed to overcome the steric hindrance through surface modification of the liposome with macromolecules such as PEG. This represents an important discovery towards the application of these new synthetic materials in biological systems.
To realise the complexity and function of biological systems, van Hest and coworkers (DOI: 10.1039/c1jm12407b) have harnessed the concept of polymersomes compartmentalisation to allow for the positional assembly of essential enzymes and the exploration of their as cofactor regenerating nanoreactors (Fig. 3). Inspired by the phase separated structure of these assemblies the authors have for the first time demonstrated their potential as an efficient cofactor recycling system for application in cell-free Baeyer–Villiger enzyme mediated catalysis.
 | ||
| Fig. 3 Schematic highlighting the application of polymersomes as biomimetic scaffolds for the regeneration of cofactor NADP+ inside the polymersome, allowing for efficient Baeyer–Villiger reaction. | ||
Concluding remarks
Synthetic biology offers a host of exciting scientific opportunities, and may have profound societal and economic consequences. Ethical issues have informed the debate on synthetic biology from the start, and public dialogue initiatives combined with ‘citizen science’ are shaping the development of potential practical applications. Harnessing the power of biological machinery has been theoretically possible since the advent of gene technologies but synthetic biology offers the chance to adapt biology into wholly new areas. Materials chemistry is a vital part of these developments, as control over molecules at the length scale of cells is the key to directing synthetic cell function.4 Importantly, any practical application will require molecular precision built in from the start, as reproducibility and reliability are fundamental attributes of any technology. In addition, for applications in the biomedical field such as in vivo smart sensors or drug delivery vehicles, chemistries need to be robust and fully understood in complex environments. Examples might include synthetic gene circuits, which in a sensor would need to detect a substrate in low concentrations then amplify the signal with absolute fidelity every time over a long time period, or responsive release polymer constructs, which would need to alter conformation in exactly predictable ways when triggered by a biological event. We do not yet have such control over these types of materials chemistry, but the papers sampled in this editorial indicate that many of the underlying synthetic and technical issues are being addressed. If progress continues to accelerate, ultimately one can envisage robust and scalable chemistries being used to generate protocells and synthetic organelles with functions ranging from catalysis, diagnostics and drug delivery through perhaps even to replication and evolution.5Cameron Alexander, University of Nottingham, UK and Rachel O’Reilly, University of Warwick, UK.
References
- D. G. Gibson, J. I. Glass, C. Lartigue, V. N. Noskov, R. Y. Chuang, M. A. Algire, G. A. Benders, M. G. Montague, L. Ma, M. M. Moodie, C. Merryman, S. Vashee, R. Krishnakumar, N. Assad-Garcia, C. Andrews-Pfannkoch, E. A. Denisova, L. Young, Z. Q. Qi, T. H. Segall-Shapiro, C. H. Calvey, P. P. Parmar, C. A. Hutchison, H. O. Smith and J. C. Venter, Science, 2010, 329, 52–56 CrossRef CAS.
- S. S. Mansy, J. P. Schrum, M. Krishnamurthy, S. Tobe, D. A. Treco and J. W. Szostak, Nature, 2008, 454, 122–U110 CrossRef CAS.
- A. L. Boyle and D. N. Woolfson, Chem. Soc. Rev., 2011, 40, 4295–4306 RSC.
- G. Pasparakis, N. Krasnogor, L. Cronin, B. G. Davis and C. Alexander, Chem. Soc. Rev., 2010, 39, 286–300 RSC.
- L. Cronin, N. Krasnogor, B. G. Davis, C. Alexander, N. Robertson, J. H. G. Steinke, S. L. M. Schroeder, A. N. Khlobystov, G. Cooper, P. M. Gardner, P. Siepmann, B. J. Whitaker and D. Marsh, Nat. Biotechnol., 2006, 24, 1203–1206 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2011 |

