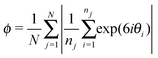Open Access Article
Open Access ArticleDynamic ordering of nuclei in syncytial embryos: a quantitative analysis of the role of cytoskeletal networks†
Takuma
Kanesaki‡
a,
Carina M.
Edwards‡
b,
Ulrich S.
Schwarz
*bc and
Jörg
Grosshans
*a
aInstitute for Biochemistry, Medical School, University of Göttingen, 37077 Göttingen, Germany. E-mail: jgrossh@gwdg.de; Fax: +49-551-39-14614; Tel: +49-551-39-8242
bBioquant, University of Heidelberg, 69120 Heidelberg, Germany. E-mail: ulrich.schwarz@bioquant.uni-heidelberg.de; Fax: +49-6221-54-9331; Tel: +49-6221-54-9399
cInstitute for Theoretical Physics, University of Heidelberg, 69120 Heidelberg, Germany
First published on 14th October 2011
Abstract
In syncytial embryos nuclei undergo cycles of division and rearrangement within a common cytoplasm. It is presently unclear to what degree and how the nuclear array maintains positional order in the face of rapid cell divisions. Here we establish a quantitative assay, based on image processing, for analysing the dynamics of the nuclear array. By tracking nuclear trajectories in Drosophila melanogaster embryos, we are able to define and evaluate local and time-dependent measures for the level of geometrical order in the array. We find that after division, order is re-established in a biphasic manner, indicating the competition of different ordering processes. Using mutants and drug injections, we show that the order of the nuclear array depends on cytoskeletal networks organised by centrosomes. While both f-actin and microtubules are required for re-establishing order after mitosis, only f-actin is required to maintain the stability of this arrangement. Furthermore, f-actin function relies on myosin-independent non-contractile filaments that suppress individual nuclear mobility, whereas microtubules promote mobility and attract adjacent nuclei. Actin caps are shown to act to prevent nuclear incorporation into adjacent microtubule baskets. Our data demonstrate that two principal ordering mechanisms thus simultaneously contribute: (1) a passive crowding mechanism in which nuclei and actin caps act as spacers and (2) an active self-organisation mechanism based on a microtubule network.
Insight, innovation, integrationThe application of quantitative analysis to tissue development and morphogenesis is providing new and important insights into the underlying mechanisms. Here we use automatic image processing to follow changes in the spatial organisation of nuclear positioning in the early (syncytial) insect embryo throughout the cell cycle. We combine this analysis with a range of drug-injection experiments and mutations. By defining appropriate mathematical measures for the degree of system order, we are able to draw conclusions about the differing roles of the cytoskeletal elements, and the mechanisms by which they drive the ordering process. Thus the integration of quantitative image processing, statistical data analysis and experimental investigation enables us to derive significant insights into tissue organisation in a paradigm system. |
1 Introduction
Many types of epithelia are characterized by a hexagonal arrangement of cells.1,2 This arrangement is usually driven by both inter-cellular adhesion and intracellular forces generated by the cytoskeleton, which act over multiple time- and length-scales. The dominant effect in this organizational process is the large tension in the cell membranes, which makes the system foam-like, leading to a preference for hexagonal ordering to minimize the interfacial energy.3,4 As the epithelial layer grows, the cell rearrangements are determined mainly by these tensional forces. It has been recently argued that cell migration in such layers shares many similarities with the slow dynamics in glass-forming systems as cell movement becomes progressively slower and more collective with increasing density.5 An important aspect of this and other similar studies is the use of quantitative image analysis of the experimental data to suggest physical mechanisms underlying the biological phenomena. Indeed during recent years, it has been demonstrated that such quantitative analysis can enable us to infer many important conclusions about the morphogenesis of epithelia6,7 and the regulation of tissue size.8Although epithelial tissues are a rewarding model system for quantitative analysis of tissue growth, it is worth noting that other model systems exist which are even more simple in this regard. In contrast to epithelia, syncytial insect embryos lack membranes separating the nuclei into cells, so wall tension does not dominate the dynamics in this system. Syncytial embryos go through a rapid sequence of cell divisions and after each division, the nuclei are observed to rapidly self-organise over a time-scale of only a few minutes. The main forces driving this process are assumed to be cytoskeletal in origin, with each nucleus in the array being associated with an actin cap, a pair of centrosomes and an enclosing microtubule basket.9,10 These elements change dynamically, altering their arrangement, size and function as the nuclei pass through the various stages of the cell cycle. The nuclear rearrangements, and the subsequent maintenance of positioning, are thus generated within a rapidly changing structural network.11 Here, we use quantitative image analysis to investigate this process in the popular developmental system of Drosophila melanogaster. In contrast to the case of epithelial tissues, our setup enables direct observation of the underlying cellular mechanisms.
Early Drosophila development involves the proliferation of nuclei through 13 rounds of consecutive mitoses without cytokinesis.9,12 During the first 9 cycles the nuclei synchronously divide in the interior of the embryo. Most of the nuclei then move outwards and uniformly distribute at the cortex to form a nuclear array. The last four nuclear divisions start at both anterior and posterior poles and propagate to the centre within about a minute in a wave-like manner. During these mitoses the nuclei significantly move and the array is severely perturbed. Generation of regularity in the array after mitosis is observed in this system, a process assumed to be essential for determining a uniform cell size. In particular, a uniform cell size and regular cellular packing have been hypothesised to be important for establishing precise morphogen gradients,13 and for the establishment of planar cell polarity signalling in later development.14
Despite previous studies of cytoskeletal organisation in the early Drosophila embryo,9,10,15,16 the dynamics of nuclear ordering throughout the cell cycle and an explanation for how the cytoskeleton mechanically controls this ordering have not been established. In the absence of inter-cell membranes, it is possible for large-scale networks composed of microtubules and f-actin networks to span the embryo. However, this leaves unanswered whether and to what degree this network is mechanically active and directly responsible for the ordering process. Due to the rapidity of development, and the near-synchrony of nuclear division, it is possible to observe the dynamical rearrangement of the nuclei over complete cycles. Using this advantage we develop a quantitative assay based on tracking the movements of the nuclei. We evaluate order parameters describing the regularity of the nuclear array and follow these through the cell cycle. In combination with mutant and drug-injected embryos, we use this assay to look at the individual roles of f-actin and microtubules in driving the emergence of order in this minimal system.
2 Methods
2.1 Genetics
Nuclei in wildtype and mutant embryos were labelled by expression of a Histone H2Av-GFP/RFP construct17 or injection of Alexa488-labelled Histone1 (Invitrogen). The following mutants and transgenes were used: Map60[KG00506], fs(1)maternal haploid, P{spq-GFP/RFP-moesin, w[+]},18 P{UASp-eb1-GFP, w[+]},19 P{UASp-tubulin-GFP, w[+]} both driven by P{tub-Gal4-VP16, w[+]}, and P{Ubq-GFP-Dsas6}.20 If not otherwise noted, fly stocks were obtained from the Bloomington stock centre. P{H2Av-mRFP, w[+]} was cloned by inserting mRFP21 instead of GFP into a Histone H2Av genomic construct.172.2 Time-lapse imaging
Dechorionated embryos were fixed on a coverslip and covered with halocarbon oil. Time lapse recordings were performed with an inverted microscope (Carl Zeiss) with UltraVIEW ERS spinning disk optics (Perkin Elmar) and objectives EC Plan-NEOFLUAR 40x/1.3/oil and Plan-APOCHRMAT 100x/1.4/oil. The temperature was 20–23 °C. We took care to minimise the exposure of the embryo to light and mechanical disturbance, both of which may induce a nuclear fall-out phenotype and abnormal behaviour of both the nuclei and mitotic waves. The recordings usually lasted 10–30 min, comprising one cell cycle with a frame rate of 0.1/s. For the Histone H2Av-GFP/RFP channel three to four z sections covering 6–8 μm were merged.2.3 Microinjection of drugs
100 μg ml−1colcemid, 500 μg ml−1latrunculin A, or deionized water were injected into embryos expressing Histone H2Av-GFP/RFP and Tubulin-GFP/Moesin-GFP/RFP. The embryos were devitellinized, slightly desiccated, and aligned on a cover-slip. Developmental stages of embryos were corroborated by imaging of nuclei, and each toxin solution of the toxin was injected on the spinning disk microscope at the desired time. The time-lapse recordings were interrupted for 30–60 s during the injection. 100 μg ml−1aphidicolin was injected into preblastoderm stages of embryos. Afterwards the embryos were incubated for 1 h at room temperature and time-lapse imaging performed.2.4 Immunostaining of embryos
Embryos expressing SAS6-GFP were dechorionated with hypochlorite (50% bleach), fixed in a mixture of 37% formaldehyde and heptane for 30 min, and manually devitellinized. The fixed embryos were rinsed with phosphate-buffered saline (PBS, 12 mM sodium phosphate [pH 7.4], 137 mM NaCl, 2.7 mM KCl) containing 0.2% Tween 20, incubated for 1 h in blocking solution containing PBS with 0.5% TritonX100 and 5% bovine serum albumin, and treated with alpha-Tubulin antibody (0.7 μg ml−1; B512 clone, Sigma) at 4 °C overnight. After washing steps, the embryos were stained with Alexa546-labelled secondary antibody (5 μg ml−1, Molecular Probes), Alexa647-labelled phalloidin (5 μg ml−1, Molecular Probes) and 4′,6-diamidin-2-phenylindole (DAPI, 0.2 μg ml−1, AppliChem). The stained embryos were mounted in Aquapolymount (Polyscience) and photographed with a confocal microscope (Leica SP2). SAS6-GFP was imaged by its native fluorescence.2.5 Image processing
All image analysis was carried out using MATLAB (The MathWorks, Natick, MA). Image quality was improved using background subtraction and Wiener filtering. Images were segmented using Otsu's method (implemented in the MATLAB function ‘graythresh’), with morphological opening and closing performed as necessary to remove artifacts and smooth nuclear edges. Where large variations in fluorescence were observed, those regions exhibiting higher fluorescence were segmented separately. Touching cells were resolved using marker-based watershed segmentation with additional hand adjustment where necessary. For the data analysis only points away from the image edges were considered. For haploid embryos, the regions were adjusted as necessary to remove deformed or fused nuclei. A small number of time points were discarded where this effect dominated the data. In the injection experiments the region of interest was restricted to that affected by the drug, and when necessary adjusted to exclude fused (as opposed to touching) nuclei that could not be accurately segmented.Nuclear tracking was performed on segmented images using a custom routine based on minimising nuclear displacements between frames, with tracks checked by eye. A degree of coordinated nuclear movement was observed within some embryos subsequent to mitosis possibly attributable to cytoplasmic flow; those tracks displayed in Fig. 6 were selected on the basis of being those in which this feature was least prominent to facilitate comparison. To calculate the maximum separation of nuclei in anaphase, daughter pairs were identified by eye and the tracking routine applied to each pair up until the end of anaphase.
2.6 Data analysis
The neighbours of each nucleus were identified using Voronoi tessellation. The state of the system at each time point was then determined by calculating the distance d between the centre points of each pair of neighbours, and the orientation of pairs of neighbours quantified by calculating for each nucleus the angles θi that its neighbours made to the vertical. In order to determine the degree of order in the system we calculated two measures from this data: σ/μ, the standard deviation normalized by the mean internuclear distance μ; and ϕ, an averaged local orientational order parameter.The standard deviation σ captures the degree of regularity in the spacing of points. Orientational order parameters describe how closely the distribution of points approximates the orientation expected in an ideal lattice, i.e. a hexagonal array. A common measure of orientational order is the bond orientational order parameter see, for example.22,23 We choose, however, to consider the averaged local orientational order parameter,24
Mutant and wildtype data were aligned using the time of nuclear division, as identified with the point of maximum increase in the number density. This was calculated by fitting the data with a hyperbolic tangent function. (An exception was a single haploid data set that did not include a mitosis.) In Fig. 1D and E the time axis was rescaled so that the points of maximum disorder all occurred at the same time, with the division time fixed at 4.1 min. This rescaling accounted for the slight variation in the time taken for maximum disorder to be achieved and highlighted the common features of the curves; the unscaled curves are reproduced in Fig. S1A, B ESI.† The injection experiments were aligned relative to the time of injection. The gap in these curves indicates only the point of injection and does not represent the time taken.
 | ||
| Fig. 1 Quantitative analysis of the nuclear array. (A) Schematic drawing of the ellipsoidal embryo with inset showing the cortical organisation of the nuclei, microtubules and f-actin. (B) Sample image of the nuclear array in a wildtype embryo. The left half shows the fluorescence image of the embryo expressing Histone H2Av-GFP in interphase 14. The right half shows the segmentation of the array. Scale bar 20 μm. Colour coding for number of nearest neighbours as indicated. (C) Time-course, with colour-coding for individual embryos, of the nuclear density ND. (D) Proportion of five-, six-, seven-fold neighbourhood arrangements P(n) (shading indicates s.e.m, n = 16). | ||
Errors given are standard errors in the mean, calculated as detailed in ref. 25; the standard error in the sample standard deviation was taken as σ/(2n − 2)1/2, with n the sample size. The error in the time of maximum separation and maximum disorder was taken as 5 s, i.e. half the interval between images.
3 Results
In the absence of separating cell membranes the prime candidates for generating and transmitting forces in the embryo are f-actin, which builds up a cortical layer and forms the actin caps, and the microtubule network, which forms a basket around each nucleus (Fig. 1A).10,15 In addition to the nuclei-associated microtubule baskets, there exist astral microtubules beneath the outer surface membrane enclosing the egg, which point in all directions and overlap with the asters originating from centrosomes of neighbouring nuclei. To investigate the role of these elements in nuclear positioning, we have established an assay to quantify the nuclear dynamics focusing on measuring the degree of order in the system. Following time-lapse confocal microscopy of living embryos with fluorescence-labelled chromosomes, the time-course of nuclear coordinates was extracted by image processing (Fig. 1B, Movie 1, Movie 2 ESI†). We used the time series of the nuclear number density to identify the exact time point of division (Fig. 1C).We first examined the number of neighbours for each nucleus. We found that 56.1 ± 0.4% have six neighbours (mean ± standard error over 16 embryos from minutes 9–13 in 14th interphase), while 22.1 ± 0.2% and 19.5 ± 0.2% have five and seven neighbours, respectively. In contrast to the situation in Drosophila wing discs,26 in which the proportion of hexagons continually increases from about 40% to 80%, we found that these proportions remain relatively constant during interphase (Fig. 1D). The slight asymmetry between five and seven neighbours observed here is smaller than that normally observed in epithelial tissue.
Although the proportion of nuclei with six neighbours gives us a first measure of the degree of regularity, more detailed measures can be defined. We quantified the regularity of the array by calculating two variables as a function of time: (1) the standard deviation of nearest neighbour distances σ, which measures order in spatial distance, and (2) an averaged local orientational order parameter ϕ, which measures the degree of hexagonal order in the system and is calculated from the angles the nearest neighbours make to the vertical (Fig. 2A). Here σ is normalized by the average distance μ in order to be able to compare situations with different number densities. The higher the degree of order in the system the higher or lower the values of ϕ and σ, respectively.
 | ||
| Fig. 2 Ordering of the nuclear array. (A) Schematic showing the distances d, and angles θ, from which the order parameters σ/μ and ϕ are calculated. The definition of ϕ is also shown. (B, C) Time courses of σ/μ (B) and ϕ (C) before and after mitosis 13, with colour-coding for individual embryos. (D) Nuclei in a wildtype (WT) embryo in interphase 14 and a maternal haploid (mh) embryo in interphase 15 are labeled with Histone H2Av-GFP and injected Alexa488-Histone H1, respectively. Scale bar 10 μm. (E, F) Time course of σ/μ (E) and ϕ (F) in a representative wildtype embryo (grey), and mh embryos (coloured markers) in interphase 15. | ||
In general both parameters showed a characteristic behaviour during the cell cycle (Fig. 2B and C): (1) a peak of σ and a drop of ϕ indicating the disruptive effect of mitosis, followed by a recovery of order composed of (2) a rapid increase in order at the exit from mitosis and a subsequent (3) slower increase in the following interphase. The time course of σ actually resolves a double peak with the first smaller one corresponding to the onset of chromosome segregation and the introduction of small distances. After mitosis, we see that the order of the nuclear array is re-established in a biphasic manner after its rapid perturbation by near-synchronous nuclear duplication.
In inert physical systems like colloidal suspensions, increasing the particle density will drive an increase in system ordering.27 To examine the effect of a high number density on the nuclear array, we took advantage of haploid embryos that undergo an extra (14th) nuclear division28 (Fig. 2D and F). The extracted data for σ and ϕ show that order is not increased compared to wildtype (Fig. 2E and F). At lower nuclear densities in cycles 11–13 the order curves showed a stronger embryo to embryo variation (data not shown), suggesting that a certain level of number density is required for robust regulation of ordering. Taken together, these results show that ordering of the nuclear array in interphase 14 is not solely caused by a crowding effect, suggesting that other interactions may also be involved.
Having established the quantitative assay we first investigated the process of disordering the array during mitosis. There are two possibilities. The nuclear array may be disordered purely as a result of the introduction of small inter-cell distances into the system at nuclear division. Alternatively, the nuclear array may be disordered by the active elongation of the spindle that pushes the daughter chromosomes apart. By tracking the separation of daughter nuclei, we found that the time of maximal disorder (measured by both σ and ϕ) correlates with the time of maximal separation (Fig. 3A). In order to further confirm that anaphase spindle elongation drives disordering, we employed a mutant with shorter anaphase spindle elongation. We have observed that in Map60 embryos29,30 the anaphase spindle is shortened to 11.5 ± 0.6 μm, compared to 13.7 ± 0.8 μm in wildtype (Fig. 3B, n(wildtype) = 27, n(Map60) = 30 spindles, p-value < 10−13). Measuring the order of the nuclear array in these mutants, we found that the typical sharp peak/drop of σ and ϕ in mitosis is less pronounced as compared to wildtype embryos (Fig. 3C and D). For both mutant and wildtype, the normalized maximal separation correlates strongly with maximal disorder as quantified by σ (Fig. 3E). Thus we conclude that the disordering process finishes around the time at which maximal separation is achieved. If doubling of nuclear density were to be the cause of disordering, no such correlations would be expected. This suggests that the active elongation of the spindle is the cause of disordering in the nuclear array.
 | ||
| Fig. 3 Disordering in mitosis is driven by spindle elongation. (A) Temporal correlation of maximal separation of daughter chromosomes (Time Max Sep) and maximal disorder (Time Max Disorder) as measured by σ/μ and ϕ in wildtype embryos. Line shows Time Max Sep = Time Max Disorder, and is given as a guide to the eye. (B) Fluorescence images of wildtype and Map60 embryos with Histone H2Av-GFP in anaphase 13. Arrows show the distance between daughter nuclei. Scale bar 10 μm. σ/μ (C) and ϕ (D) in wildtype (dashed lines, grey) and Map60 (lines in green) embryos. (E) Correlation of the average maximal distance between daughter chromosomes normalized by average inter-nuclei distance (Norm Max Sep) and the maximal disorder as measured by σ/μ in 16 wildtype (blue) and 6 Map60 embryos (green). | ||
Before investigating further the functional importance of the cytoskeletal network to ordering we first comment on its organisation and relation to nuclear positioning. One possibility is that the nuclei directly structure the associated cytoskeleton, cytoplasm and cortical membrane themselves. Alternatively, the nuclei could be passive markers for the network, with the centrosomes as the organising component.31 To demonstrate that nuclei arrange according to the positions of centrosomes, we generated embryos deprived of almost all nuclei, but containing proliferating centrosomes, by inhibiting S phase by aphidicolin injection.31 As in normal embryos the centrosomes migrated to the cortex in cycle 9 and formed an array (Fig. S2 D,E ESI†). Fluorescent labelling of the cytoskeleton showed a network of overlapping asters and actin caps associated with the centrosomes. This observation confirms that the centrosomes are able to induce, and participate in, the cytoskeletal network and may act to organise the nuclear array. However, we note that in embryos where we introduce extra centrosomes not associated with nuclei,20 these tended to cluster and occupy nuclear free spaces (Fig. S2 A–C ESI†), suggesting that nuclear volume exclusion does contribute to some degree to the array.
To test the respective functions of the cytoskeletal networks in the ordering of the nuclear array, we injected colcemid or latrunculin to disrupt microtubules or f-actin networks, respectively.9,32 As we observed a biphasic recovery of order after mitosis we injected the drugs both immediately after anaphase in mitosis 13 (early injection) and at the onset of interphase about 5 min later (late injection) (Fig. 4A). An experimental complication is that f-actin links the nuclei to the cortex at the onset of interphase. About half of the cortical nuclei were lost when latrunculin was injected at the end of anaphase but not when injected at the onset of interphase. The efficiency of the injection was controlled in embryos expressing the f-actin marker moesin-GFP and Histone-RFP.
 | ||
| Fig. 4 Investigating the function of microtubules and f-actin. (A) Timeline of the drug treatment. Water, colcemid or latrunculin A was injected into the embryos either at the end of anaphase (1–2 min after onset of the anaphase, early injection) or at the onset of the interphase (4–6 min after the onset of the anaphase, late injection). (B, C) Time course of order parameters σ/μ (B) and ϕ (C) after early injection. The gap in the graphs indicates point of injection, and lines correspond to individual embryos. (D, E) The order parameters σ/μ (D) and ϕ (E) after late injection, the gap in the curves indicates time of injection. (Shading indicates s.e.m, n = 4 colcemid/latrunculin, n = 3H2O.) | ||
Injecting latrunculin A immediately after anaphase we observed two prominent features indicating a severely perturbed nuclear array: touching nuclei and large internuclear gaps. The order as measured by σ and ϕ was lower in latrunculin A-injected embryos than in control embryos (Fig. 4B and C). The ϕ profile appeared erratic and did not reach a stable plateau in phase 2. The perturbed nuclear array is not simply due to the lower nuclear density, as can be seen by comparison to wildtype embryos in interphase 13. We cannot exclude that the gaps are due to the lost nuclei, but it is unlikely that the touching nuclei are a consequence of the reduced nuclear density. Similarly for late injection, which disrupted f-actin when the nuclear array was already ordered, a similar phenotype of touching nuclei and large gaps was observed and a consequent drop in order measured (Fig. 4D and E). These data show that f-actin is required for both establishing and maintaining the ordered nuclear array.
The colcemid injected embryos were used to test the function of microtubules in ordering the nuclear array, using both early and late injection according to the protocol of latrunculin injection. We observed in the early injected embryos that ϕ was not immediately recovered and only showed a slow increase afterwards, demonstrating an essential function for microtubules in establishing the ordered nuclear array (Fig. 4B and C). However, colcemid injection in the later maintenance phase did not obviously perturb the nuclear array (Fig. 4D and E). In this latter case both σ and ϕ remained at levels commensurate with the values of water injected embryos. Thus in contrast to f-actin, microtubules are essential for establishing but not for maintaining the ordered nuclear array.
An interesting question is whether the f-actin network or the microtubules are passive or active generators of force. F-actin, for example, could together with myosin generate active contractile forces in the embryo. In the interphases of the blastoderm stage distinct patterns of myosin II and f-actin are observed,33,34 with myosin excluded from the f-actin caps and uniformly distributed in the area in between. Although Myosin is clearly involved in the distribution of the nuclei along the embryonic axis before cortical migration,35 its roles during the cortical cycles are less clear. We tested whether myosin is involved in nuclear arrangement by inhibiting Rho kinase, which activates Myosinviaphosphorylation of the myosin regulatory light chain (spagetti-squash (spq) in Drosophila). We employed the effective Rho kinase inhibitor Y-27632 and controlled its activity in embryos expressing spq-GFP.36,37 In such embryos we recorded the nuclear dynamics and measured the order parameters (Fig. 5A and B). Since no obvious difference to the nuclear dynamics of wild-type embryos was observed, myosin II activity and Rho kinase signaling seem not to be essential for nuclear interactions. These experiments support the model that f-actin acts mostly through non-contractile filaments and does not rely on generating active forces.
 | ||
| Fig. 5 Testing for f-actin and microtubule generated active forces. (A,B) Rho kinase and MyoII are not required for an ordered nuclear array. σ/μ (A) and ϕ (B) of the Y-27632-injected embryos (green lines), against wildtype embryos (grey dashed lines). (C-E) Microtubule asters attract neighbouring nuclei in the absence of f-actin. (C, D) Fluorescence images of embryos expressing EB1-GFP after early injection (interphase 14) with latrunculin A (C) or latrunculin A and colcimid (D). The images were recorded 5 min after injection. Each image is composed of two confocal sections at different depths (apical, green; basal, red) as indicated in the schematic. Arrows indicate centrosomes that have moved in between multiple nuclei. Scale bar 10 μm. (E) Fraction of touching pairs of nuclei (frequency) that have a centrosome at their interface. The experiments were performed for two injected embryos each (X-axis). For each embryo at least 30 nuclear pairs were scored. | ||
Considering how the microtubules are involved in nuclear interactions we see that astral microtubules originating from adjacent centrosome pairs could align in anti-parallel orientation and generate a force by pushing or pulling as observed in the mitotic spindle.38,39 We visualised the microtubule dynamics in the embryo by labelling the growing (+) ends with EB1-GFP (Movie 3 ESI†). As soon as the nuclei with their centrosomes have reached the cortex, microtubule asters are observed. Time-lapse recordings of such embryos showed that the growing tips often extend to areas of adjacent asters. They are observed to frequently intermingle and to form potential anti-parallel pairings that may potentially be involved in force generation (Fig. S3 ESI†).
The microtubules could also interact directly with the neighbouring nuclei in addition to their own nucleus. To test this hypothesis that microtubules may interact directly with adjacent nuclei, we investigated microtubule organisation and nuclear behaviour in embryos deprived of f-actin. Latrunculin was injected into embryos expressing EB1-GFP. In these embryos we observed with a high frequency that centrosomes move between two, three or even four nuclei, something which was not observed in wild-type embryos (Fig. 5C and E, Movie 4 ESI†). This behaviour demonstrates that microtubule asters can attract adjacent nuclei. That this phenotype depends on the action of microtubules is clear as this behaviour was not observed in embryos coinjected with latrunculin and colcemid (Fig. 5D and E). These data show that microtubules can attract adjacent nuclei and that f-actin normally restricts this attractive activity.
Finally, we note that the non-contractile f-actin could still act passively to stabilise and counteract the force-generating network of microtubules. We observed when injecting latrunculin A into the embryos a greater mobility of nuclei (Movie 4 ESI†) than in wild-type embryos. In Fig. 6A–D we plot the traces of the centre of mass of the nuclei over a period of 2.7 min. The higher mobility is a specific effect requiring the function of microtubules, since the injection of both latrunculin and colcemid lead to immobile nuclei (Fig. 6D). The low nuclear mobility in wild-type embryos was not affected by injection of colcemid alone (Fig. 6B). These data indicate that f-actin could have a role in stabilising the nuclear array by dampening microtubule dependent nuclear mobility.
 | ||
| Fig. 6 F-actin dampens microtubule dependent nuclear movement. (A–D) Traces of nuclear movement over a period of 2.7 min after late injection of deionized water (A), colcemid (B), latrunculin A (C), and latrunculin A and colcemid simultaneously (D). Scale bar 20 μm. | ||
4 Discussion
In contrast to epithelia, in which cells interact by a combination of extra- and intracellular forces,4,26,40–45 interactions of syncytial nuclei involve only cytoplasmic components and intracellular forces. Within the embryo the lack of separating cell membranes means that the nuclear array can be treated as a two-dimensional network of individual nuclei that are locally strongly cross-linked. Furthermore, interactions and rearrangements of the syncytial nuclei proceed rapidly over a time-scale of seconds to minutes whereas epithelia develop over time-scales of up to days. These properties of the nuclear array in syncytial embryos makes it an ideal system for investigating the self-organization of a global network through local interactions.In this study, we have focused on the interaction of nuclei in the paradigm system of the syncytial blastoderm of Drosophila melanogaster. We have established a quantitative assay for measuring temporal and spatial changes in the nuclear array, which we have used to study the mechanisms underlying network formation and maintenance. For reasons of simplicity we restricted our recordings to a single field of view comprising up to a few hundred of the 6000 nuclei. Although the complete network including the highly bended poles could be recorded by suitable microscopy such as Selective Plane Illumination Microscopy,46 together with appropriate representations of the nuclear array,47 the principal observations and conclusions concerning network formation would probably not be much different. Consistent with previous qualitative observations,9,10,48 our quantitative measurements show that the nuclear array switches between unordered and ordered states depending on the cell cycle stage. Our quantitative assay allows us to track the dynamics of this ordering process. We find that at the onset of anaphase order is rapidly destroyed in the array but that this is quickly followed by a biphasic phase consisting of a short rapid recovery followed by a second slower phase of rearrangement. Furthermore we have shown by tracking pairs of daughter nuclei that disordering is primarily caused by spindle elongation during anaphase and not by the initial doubling and subsequent placement of new nuclei within the array.
Drug injections as well as analysis of mutants have revealed essential and distinct functions of both microtubules and f-actin in establishing and maintaining order in the nuclear array and in controlling nuclear mobility. We found that actin is essential for both the initial fast phase of recovery of order and also in the second phase of dynamics in which order is gradually increased and maintained. In contrast, we found that the microtubule network primarily affects the initial phase, and is not essential for maintaining order in the array. As both the microtubule network and the actin caps grow in the interphase period after mitosis, recovering their structure after the dissolution of both during mitosis, differences in the growth rates of these two elements could be a contributing factor to the biphasic nature of the recovery.
We investigated the possible mechanisms by which actin and microtubules could generate the observed dynamic ordering. The actin caps that are linked with each of the nuclei may act as passive spacers or bumpers for the nuclei, restricting individual nuclear movement of nuclei and shielding nuclei from the attraction of neighbouring microtubule asters. However, we note that we also saw that at higher nuclear densities in haploid embryos there is no additional ordering of the array. The growth of the actin caps during interphase15 may allow repositioning during the initial phase of ordering, which then stabilise the array when the actin caps have expanded until they touch each other. The function of actin seems mainly to be based on non-contractile filaments, since we could not detect any contribution to ordering from myosin activity. F-actin may also be indirectly involved in neighbour interactions by controlling the viscoelastic properties of the cytoplasm,49 further acting to dampen nuclear movements.
In contrast, microtubules may play an active role in generating attractive or repulsive forces. Microtubules of asters originating from adjacent centrosomes and nuclei are observed to directly interact, which could generate forces by the action of motors or crosslinkers, such as kinesin-5.50,51 Specifically, we observed that microtubules from adjacent asters form antiparallel pairs, that are not observed in normal interphase cells and that these could be the site of force generation as in the mitotic spindle. We also observed an attractive interaction of microtubles with adjacent nuclei in the absence of f-actin, in that multiple nuclei are incorporated in a microtubule aster originating from a single pair of centrosomes. Microtubules may interact with the overlaying actin caps by dynein-dynactin complexes that are linked to the caps.52,53 Such asymmetrical interactions would center the centrosomes and its nucleus below the actin cap.15 Finally, we note that the nuclear array may be genetically controlled; depending on anterior group genes and zygotic gene expression, the nuclei in the anterior third of the embryo are more widely spaced.11
Here we provide evidence that the formation of the regular array involves active as well as passive interactions between the nuclei mediated by the cytoskeletal networks. Such an understanding presents an excellent starting point from which to investigate how ordering processes tie in with the biological function of the growing tissue, including the establishment of morphogen gradients and planar cell polarity.
Acknowledgements
We would like to thank D. Brunner, D. Kiehart, J. Raff, E. Schejter and the Bloomington stock centre for sharing fly stocks or materials. JG acknowledges support by the German Research Council and SFB 937. CME was supported by a postdoctoral grant from the Center for Modelling and Simulation in the Biosciences (BIOMS) at Heidelberg. JG and USS are members of the cluster of excellence CellNetworks at the University of Heidelberg.References
- W. M. Roberts, J. Howard and A. J. Hudspeth, Annu. Rev. Cell Biol., 1988, 63–92 CrossRef CAS.
- D. Nilsson, in Facets of vision, ed. D. Stavenga and R. Hardie, Springer Verlag, 1989, ch. Optics and evolution of the compound eye, pp. 30–73 Search PubMed.
- J. A. Glazier and D. Weaire, J. Phys.: Condens. Matter, 1992, 4, 1867–1894 CrossRef.
- R. Farhadifar, J.-C. Roper, B. Aigouy, S. Eaton and F. Jülicher, Curr. Biol., 2007, 17, 2095–2104 CrossRef CAS.
- T. E. Angelini, E. Hannezo, X. Trepat, M. Marquez, J. J. Fredberg and D. A. Weitz, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 4714–4719 CrossRef CAS.
- T. Bittig, M. Wartlick, M. González-Gaitán and F. Jülicher, Eur. Phys. J. E, 2009, 30, 93–99 CrossRef CAS.
- G. B. Blanchard, A. J. Kabla, N. Schultz, L. Butler, B. Sanson, N. Gorfinkiel, L. Mahadevan and R. J. Adams, Nat. Methods, 2009, 6, 458–464 CrossRef CAS.
- L. Hufnagel, A. A. Teleman, H. Rouault, S. M. Cohen and B. I. Shraiman, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 3835–3840 CrossRef CAS.
- V. E. Foe, G. M. Odell and B. A. Edgar, in Development of Drosophila melanogaster, ed. M. Bate and A. Martinez-Arias, Cold Spring Harbor Press, 1993, ch. Mitosis and morphogenesis in the Drosophila embryo: Point and Counterpoint, pp. 149–300 Search PubMed.
- E. D. Schejter and E. Wieschaus, Annu. Rev. Cell Biol., 1993, 9, 67–99 CrossRef CAS.
- J. T. Blankenship and E. Wieschaus, Development, 2001, 128, 5129–5138 CAS.
- M. Zalokar and I. Erk, J. Mic. Biol. Cell, 1976, 23, 97–106 Search PubMed.
- T. Bollenbach, P. Pantazis, A. Kicheva, C. Bokel, M. González-Gaitán and F. Jülicher, Development, 2008, 135, 1137–1146 CrossRef CAS.
- D. Ma, K. Amonlirdviman, R. L. Raffard, A. Abate, C. J. Tomlin and J. D. Axelrod, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 18800–18805 CrossRef CAS.
- J. Cao, J. Crest, B. Fasulo and W. Sullivan, Curr. Biol., 2010, 8, 770–776 CrossRef.
- V. Foe, C. Field and G. Odell, Development, 2000, 127, 12767–12787 Search PubMed.
- M. Clarkson and R. Saint, DNA Cell Biol., 1999, 18, 457–462 CrossRef CAS.
- D. P. Kiehart, C. G. Galbraith, K. A. Edwards, W. L. Rickoll and R. A. Montague, J. Cell Biol., 2000, 149, 471–490 CrossRef CAS.
- F. Jankovics and D. Brunner, Dev. Cell, 2006, 11, 375–385 CrossRef CAS.
- N. Peel, N. R. Stevens, R. Basto and J. W. Raff, Curr. Biol., 2007, 17, 834–843 CrossRef CAS.
- R. E. Campbell, O. Tour, A. E. Palmer, P. A. Steinbach, G. S. Baird, D. A. Zacharias and R. Y. Tsien, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 7877–7882 CrossRef CAS.
- S. Torquato and F. H. Stillinger, Rev. Mod. Phys., 2010, 82, 2633–2672 CrossRef.
- J. A. Zallen and R. Zallen, J. Phys.: Condens. Matter, 2004, 16, S5073–S5080 CrossRef CAS.
- A. R. Kansal, T. M. Truskett and S. Torquato, J. Chem. Phys., 2000, 113, 4844–4851 CrossRef CAS.
- G. Squires, Practical Physics, Cambridge University Press, 1985 Search PubMed.
- A.-K. Classen, K. I. Anderson, E. Marois and S. Eaton, Dev. Cell, 2005, 9, 805–817 CrossRef CAS.
- P. Chaikin and T. Lubensky, Principles of condensed matter physics, Cambridge University Press, 1995 Search PubMed.
- B. A. Edgar, C. P. Kiehle and G. Schubiger, Cell, 1986, 44, 365–372 CrossRef CAS.
- D. R. Kellogg, K. Oegema, J. Raff, K. Schneider and B. M. Alberts, Mol. Biol. Cell, 1995, 6, 1673–1684 CAS.
- R. D. J. Butcher, S. Chodagam, R. Basto, J. G. Wakefield, D. S. Henderson, J. W. Raff and W. G. F. Whitfield, J. Cell Sci., 2004, 117, 1191–1199 CrossRef CAS.
- J. W. Raff and D. M. Glover, Cell, 1989, 57, 611–619 CrossRef CAS.
- B. A. Edgar, G. M. Odell and G. Schubiger, Genes Dev., 1987, 1, 1226–1237 CrossRef CAS.
- A. Royou, W. Sullivan and R. Karess, J. Cell Biol., 2002, 158, 127–137 CrossRef CAS.
- A. Royou, C. Field, J. C. Sisson, W. Sullivan and R. Karess, Mol. Biol. Cell, 2004, 15, 838–850 CrossRef CAS.
- S. Wheatley, S. Kulkarni and R. Karess, Development, 1995, 121, 1937–1946 CAS.
- S. Narumiya, T. Ishizaki and M. Uehata, Methods Enzymol., 2000, 325, 273–284 CAS.
- C. Bertet, L. Sulak and T. Lecuit, Nature, 2004, 429, 667–671 CrossRef CAS.
- M. E. Tanenbaum, L. Macurek, N. Galjart and R. H. Medema, EMBO J., 2008, 27, 3235–3245 CrossRef CAS.
- F. Nédélec, J. Cell Biol., 2002, 158, 1005–1015 CrossRef.
- R. Nagpal, A. Patel and M. C. Gibson, BioEssays, 2008, 30, 260–266 CrossRef.
- M. C. Gibson, A. B. Patel, R. Nagpal and N. Perrimon, Nature, 2006, 442, 1038–1041 CrossRef CAS.
- J. Käfer, T. Hayashi, A. F. M. Marée, R. W. Carthew and F. Graner, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 18549–18554 CrossRef.
- K. P. Landsberg, R. Farhadifar, J. Ranft, D. Umetsu, T. J. Widmann, T. Bittig, A. Said, F. Jülicher and C. Dahmann, Curr. Biol., 2009, 19, 1950–1955 CrossRef CAS.
- B. Monier, A. Pelissier-Monier, A. H. Brand and B. Sanson, Nat. Cell Biol., 2010, 12, 60–65 CrossRef CAS.
- T. Aegerter-Wilmsen, A. C. Smith, A. J. Christen, C. M. Aegerter, E. Hafen and K. Basler, Development, 2010, 137, 499–506 CrossRef CAS.
- J. Huisken, J. Swoger, F. Del Bene, J. Wittbrodt and E. H. K. Stelzer, Science, 2004, 305, 1007–1009 CrossRef CAS.
- S. V. E. Keränen, C. C. Fowlkes, C. L. Luengo Hendriks, D. Sudar, D. W. Knowles, J. Malik and M. D. Biggin, GenomeBiology, 2006, 7, R124 CrossRef.
- B. Sonnenblick, in Biology of Drosophila, ed. M. Demerec, Wiley, New York, 1950, ch. The early embryology of Drosophila Search PubMed.
- B. D. Hoffman and J. C. Crocker, Annu. Rev. Biomed. Eng., 2009, 11, 259–288 CrossRef CAS.
- J. Baker, W. E. Theurkauf and G. Schubiger, J. Cell Biol., 1993, 122, 113–121 CrossRef CAS.
- J. M. Scholey, Cell Motil. Cytoskeleton, 2009, 66, 500–508 CrossRef CAS.
- J. T. Robinson, E. J. Wojcik, M. A. Sanders, M. McGrail and T. S. Hays, J. Cell Biol., 1999, 146, 597–608 CrossRef CAS.
- E. N. Cytrynbaum, P. Sommi, I. Brust-Mascher, J. M. Scholey and A. Mogilner, Mol. Biol. Cell, 2005, 16, 4967–4981 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Additional data. See DOI: 10.1039/c1ib00059d |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2011 |