Structure of modified ε-polylysine micelles and their application in improving cellular antioxidant activity of curcuminoids
Hailong
Yu
,
Ji
Li
,
Ke
Shi
and
Qingrong
Huang
*
Department of Food Science, Rutgers, The State University of New Jersey, 65 Dudley Road, New Brunswick, New Jersey 08901, USA. E-mail: qhuang@aesop.rutgers.edu; Fax: +732-932-6776; Tel: +732-932-7193
First published on 20th June 2011
Abstract
The micelle structure of octenyl succinic anhydride modified ε-polylysine (M-EPL), an anti-microbial surfactant prepared from natural peptide ε-polylysine in aqueous solution has been studied using synchrotron small-angle X-ray scattering (SAXS). Our results revealed that M-EPLs formed spherical micelles with individual size of 24–26 Å in aqueous solution which could further aggregate to form a larger dimension with averaged radius of 268–308 Å. Furthermore, M-EPL micelle was able to encapsulate curcuminoids, a group of poorly-soluble bioactive compounds from turmeric with poor oral bioavailability, and improve their water solubility. Three loading methods, including solvent evaporation, dialysis, and high-speed homogenization were compared. The results indicated that the dialysis method generated the highest loading capacity and curcuminoids water solubility. The micelle encapsulation was confirmed as there were no free curcuminoid crystals detected in the differential scanning calorimetry analysis. It was also demonstrated that M-EPL encapsulation stabilized curcuminoids against hydrolysis at pH 7.4 and the encapsulated curcuminoids showed elevated cellular antioxidant activity compared with free curcuminoids. This work suggested that M-EPL could be used as new biopolymer micelles for delivering poorly soluble drugs/phytochemicals and improving their bioactivities.
Introduction
Amphiphilic polymers have both hydrophilic and hydrophobic segments. Above the critical aggregation concentration, they self-assemble to form polymer micelles in aqueous solutions. The generated polymer micelles are able to solubilize water-insoluble compounds and function as a potential formulation platform for drug delivery.1,2Many drug candidates and bioactive phytochemicals are water insoluble, which leads to limited bioavailability. Among those compounds, curcuminoids are a group of curcumin-like compounds extracted from turmeric. They have been shown to have anti-inflammatory, anti-cancer, antioxidant and anti-microbial activities.3–6 Unfortunately, its oral bioavailability was greatly limited by its water insolubility.7
Recently, we synthesized a new amphiphilic polymer, modified ε-polylysine (M-EPL), using ε-polylysine (EPL, Fig. 1A) and octenyl succinic anhydride (OSA).8EPL is a biopolymer generated by bacteria Streptomyces albulus,9,10 and is a natural antimicrobial agent.11 After chemical modification, M-EPLs (or called OSA-g-EPLs, Fig. 1B) were amphiphilic and able to lower the surface tension of water and form polymer micelles. Meanwhile, they still retained the same antimicrobial activity of EPL, rendering them bifunctional (surface active and antimicrobial) molecules.8 However, although M-EPLs were shown to form micelles in water, the micelle structure still remains unclear.
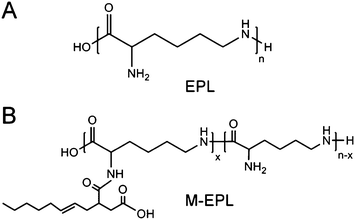 | ||
| Fig. 1 Chemical structures of epsilon polylysine (EPL) (A) and octenyl succinic anhydride modified ε- polylysine (OSA-g-EPL) (B). | ||
In this study, the micelle structures of M-EPLs were analyzed using synchrotron small-angle X-ray scattering (SAXS). Subsequently, three loading methods were compared to achieve the maximal water solubility of curcuminoids upon encapsulation in the M-EPL-based polymer micelles. Furthermore, the effects of polymer micelle encapsulation on the stability against alkaline hydrolysis and the cellular antioxidant activity of curcuminoids were also investigated.
Experimental
Reagents
Curcuminoids powder containing about 82% curcumin, 15% demethoxycurcumin (DCur) and 3% bisdemethoxycurcumin (BDCur) was received as a gift from Sabinsa Corporation (Piscataway, NJ, United States). ε-Polylyine (EPL) was purchased from Zhejiang Silver-Elephant Bioengineering Co., Ltd., China, and used without further purification. Octenyl succinic anhydride (OSA), sodium bicarbonate (NaHCO3), dimethyl sulfoxide (DMSO), and chloroform were purchased from Sigma-Aldrich. Dialysis membranes (molecular weight cutoff 1000 Da) were obtained from Spectrum Laboratories. Glacial acetic acid, HPLC-grade water and acetonitrile were from J. T. Baker.HepG2 cells were generously provided by Dr Mou-Tuan Huang from Department of Chemical Biology, Rutgers, the State University of New Jersey. Minimum Essential Medium (MEM), Hank's buffered salt solution (HBSS), RPMI-1640 media, fetal bovine serum (FBS), phosphate buffered saline (PBS), 100X penicillin and streptomycin, and 0.25% trypsin with ethylenediaminetetraacetic acid (EDTA), L-glutamine, were all purchased from Thermo Scientific. Insulin, 2′,7′-dichlorfluorescein-diacetate (DCFH-DA), 2,2′-azobis (2-amidinopropane) (ABAP), Williams' Medium E (WME), hydrocortisone were purchased from Sigma-Aldrich.
Synthesis of modified ε-polylysine (M-EPLs)
M-EPL was synthesized according to our previous paper.8 Briefly, EPL was dissolved in DMSO and different amounts of octenyl succinic anhydride were added dropwise. After 18 h reaction at about 35–40 °C, the products were dialyzed and lyophilized. Degree of substitution was calculated from 1H NMR spectra of M-EPLs. To prepare the modified EPL for curcuminoids encapsulation, 10 g EPL and 4.3 mL OSA were used.Small-angle X-ray scattering (SAXS) measurements
SAXS intensity profiles were collected at the BioCAT, 18-ID beamline, at the Advanced Photon Source, Argonne National Laboratory. The sample-detector distance was set at 2.592 m to cover a Q range of 0.006–0.3 Å−1 (with a Mar165 CCD being offset laterally relative to the X-ray beam). A flow cell of 1.5mm diameter capillary equipped with a brass block (thermostatted with a water bath) was utilized for holding samples. A MICROLAB 500 Hamilton pump was applied to load samples to the flow cell at a constant rate (10 μL s−1) during X-ray exposure to minimize radiation damage. The X-ray wavelength was 1.033 Å and a short exposure period of 1 s was used to acquire the scattering data. The whole experiment was kept at room temperature. Fifteen curves were collected for each sample and their averaged curves were utilized for further analysis. The final SAXS profiles were gained after subtracting the solvent background. Pair distribution functions were acquired to obtain the dimension parameters of modified ε-polylysine of different degrees of substitution.Encapsulation of curcuminoids into M-EPL micelles
Three loading methods were compared in the aspect of the loading capacity.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. After repeated dialysis, solutions in the dialysis bag were filtered through 0.45μm filter and freeze dried.
000. After repeated dialysis, solutions in the dialysis bag were filtered through 0.45μm filter and freeze dried.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min and then stirred overnight at room temperature. On the second day, the solution was filtered and freeze dried.12
000 rpm for 10 min and then stirred overnight at room temperature. On the second day, the solution was filtered and freeze dried.12
To quantify the curcuminoids amount encapsulated in the M-EPL micelle, lyophilized curcuminoids in M-EPL micelles were dissolved in dH2O at the concentration of 1 mg mL−1. Two volumes of HPLC-grade acetonitrile was then added in and mixed before quantification using HPLC.
In all the following assays, M-EPL encapsulated curcuminoids were obtained using the dialysis method.
Quantification of curcuminoids using high-performance liquid chromatography (HPLC)
An UltiMate 3000 HPLC system with 25D UV-VIS absorption detector (Dionex) and a Nova-Pak C18 3.9 × 150 mm column (Waters) were used. Mobile phase solvents were: (A) 2% acetic acid in HPLC-grade water, purged with helium, and (B) HPLC-grade acetonitrile. All aqueous samples were mixed with two volumes of acetonitrile and filtered through 0.22 μm filter. Fifty microlitre samples were injected into the column. Gradient elution was applied to separate the three curcuminoids: −2 to 0 min, 65% A and 35% B; 0 to 15 min, linear gradient from 35% B to 55% B; 15 to 20 min, held at 55% B; and from 20 to 21 min, B went back to 35%. Flow rate was set at 1 mL/min. Detection wavelength was fixed at 420 nm.Dynamic light scattering
M-EPL and freeze dried curcuminoids in M-EPL micelle were dissolved in dH2O at the concentration of 5 mg mL−1 and filtered through 0.45 μm filter. The particle size (hydrodynamic diameter) of the samples were then determined using dynamic light scattering method (BIC 90plus particle size analyzer, Brookhaven Instrument) at room temperature. The results were presented as mean ± standard error (n = 3).Differential scanning calorimetry (DSC)
Curcuminoids powder, curcuminoids and M-EPL mixture, and curcuminoids encapsulated in M-EPL micelle were analyzed using differentiation scanning calorimetry with a DSC823e thermal analyzer (Mettler Toledo, Columbus, OH) to detect the possible curcuminoids crystals. About 10 mg of each sample were put in the aluminum pan and the lid of the pan was penetrated to form a small hole. Samples were heated from 25 to 200 °C, at the rate of 10 °C per minute. Curcuminoids M-EPL mixture contained 5% curcuminoid powder and 95% M-EPL (w/w), and was grinded for better mixing.Hydrolysis stability of curcuminoids in M-EPL micelles
Freeze dried curcuminoids in M-EPL micelle sample were dissolved in dH2O at concentration of 1 mg mL−1 (with about 50 μg mL−1 curcuminoids). Curcuminoids in DMSO was added to dH2O to make a 50 μg mL−1 curcuminoids aqueous dispersion. Both solutions were diluted 10 times into PBS (pH 7.4). At different time intervals (15, 30, 45 and 60 min), the solutions were sampled, acidified with HCl and analyzed with HPLC to quantify the curcuminoids contents.Cellular antioxidant activity (CAA) assay for free curcuminoids and m-epl encapsulated curcuminoids
HepG2 cells (Passage 8–18) were regularly maintained in MEM with 10% FBS, 100 units/mL Penicillin and 100 μg mL−1Streptomycin. CAA assays were performed according to the literature.13 Briefly, sixty thousand cells/well were plated in a 96-well microplate in the CAA growth medium (William Medium E supplemented with 5% FBS, 10 mM Hepes, 2 mM L-glutamine, 100 units/mL penicillin 100 μg mL−1streptomycin). On the next day, cells were treated with curcuminoids, M-EPL encapsulated curcuminoids, or M-EPL alone in the treatment medium (WME + 2 mM L-glutamine + 10 mM Hepes) with DCFH-FA for 1 h. Then, cells were washed with PBS and treated with 600 μM ABAP in HBSS. Emission fluorescence intensity at 528 nm (slit size 20 nm) with excitation at 485 nm (slit size 20 nm) was recorded every 5 min for 1 h at 37 °C using a Synergy HT multi-mode microplate reader (BioTek). Cells treated with DCFH-FA and then HBSS + ABAP were used as the positive control (P.C.). Cells with DCFH-FA then only HSBB without ABAP were used as the negative control (N.C.).To calculate the CAA value of each treatment, the area-under-the-curve (AUC) for the plot of fluorescence intensity against time was calculated with trapezoidal method. Then the CAA value was calculated as
 | (1) |
Subsequently, Fa/Fu was plotted against the concentration of curcuminoids on double logarithmic scale, where Fa = CAA and Fu = 100 − CAA. EC50 was determined as the concentration where Fa/Fu = 1. By linear regression of the data on the plot, this value was able to be obtained mathematically. The molecular weight of curcumin was used to calculate the molar concentration of curcuminoids.
Statistical analysis
One way-ANOVA analysis with Holm-Sidak method was performed using the SigmaStat software.Results and discussion
Characterization of M-EPL micelles using synchrotron small angle X-ray scattering (SAXS)
In our previous studies, M-EPLs (or called OSA-g-EPLs) were shown to be able to form micelles and the critical aggregation concentrations (CAC) of M-EPLs with different degree substitution were determined.8 In the current work, the micelle structures of M-EPLs were further investigated using synchrotron small-angle X-ray scattering (SAXS). Fig. 2 displays the SAXS scattering profiles of 5 mg ml−1 OSA-g-EPLs solutions with different degrees of substitution (8.5%, 12.4%, and 20.5%, respectively). The fitting curves of those SAXS scattering profiles were simultaneously obtained from the inversed Fourier transform of pair distribution function by Irena package in Igor Pro software. The critical aggregation concentrations (CACs) for OSA-g-EPL8.5, OSA-g-EPL12.4, and OSA-g-EPL20.5 were 0.64 mg ml−1, 0.32 mg ml−1, and 0.18 mg ml−1 respectively.8 Therefore, 5 mg ml−1 was well above the CACs of the micelles and under that condition OSA-g-EPLs formed micelle aggregates. It was found that at the low Q region (Q < 0.2 Å−1), OSA-g-EPLs formed large aggregates due to the hydrophobic interaction from octenyl succinic anhydride (OSA) groups. In the medium Q region (0.02–0.1 Å−1), all of the SAXS profiles showed a broad peak at Q ∼ 0.04 Å−1, which corresponded to the spacing between micelles. The intensities and the peak positions of the above peaks depended on the micelle dimensions and the surface charge density of the micelle. In order to obtain the shape and size information of M-EPLs, pair distribution function (PDF) curves were obtained from both Irena Package, a tool imbedded in Igor Pro software provided by BIOCAT beamline of Advanced Photon Sources (APS)14 and GNOM.15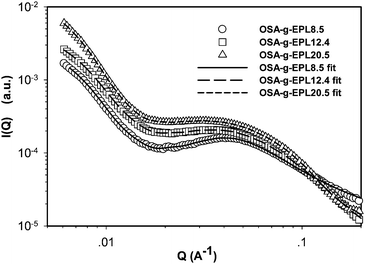 | ||
| Fig. 2 Small-angle X-ray scattering profiles of modified ε-polylysine with different degrees of substitution. The solid line (OSA-g-EPL8.5), long dash line (OSA-g-EPL12.4), and short dash line (OSA-g-EPL20.5) are fitted curves obtained from inversed Fourier transform of pair distribution function (PDF) by Irena package in Igor Pro software. PDF curves were normalized for better comparison. | ||
Fig. 3 shows the two PDF curves of OSA-g-EPL12.4 solution generated from GNOM and Irena package. It is clear that the data obtained from GNOM and Irena package have negligible difference. Therefore, only one set of dimensional parameters from Irena package were selected herein for display. As shown in Fig. 4, the bell shape of all three PDF curves indicated the spherical shape of the large micelle aggregates. However, the dimension parameters of these three OSA-g-EPLs were not identical. Table 1 lists the dimension parameters of the micelle aggregates, including maximum dimension (Dmax), radius of gyration (Rg), peak 1&2 positions and micelle number within a single aggregate. It was found that Dmax, the largest dimension within size distribution, and Rg, radius of gyration, reached the lowest value for OSA-g-EPL12.4 are 752 Å and 268 Å, respectively. The dimensions of those three OSA-g-EPLs were similar with each other although their PDF curves were not exactly overlapping. There appeared two peaks in all three PDF curves of OSA-g-EPLs. From Table 1, the first peak located at ∼24 Å, was reasonably relevant to the size of individual micelle. The second peak, located at ∼330 Å with a 15–40 Å shift for large aggregates, corresponded to the average size of OSA-g-EPLs micelle aggregate. Therefore, it was thought that the large aggregation was composed of small global micelles tightly connected with each other within the large global micelle aggregate. The aggregation number naggregation of micelles in one aggregate can be calculated through the forward scattering I (Q = 0), which was reasonably used in other amphiphilic structures as well.16 The forward scattering I (Q = 0) was obtained through a Guinier fit of the scattering profile in the low Q range. It was shown that the aggregation number within one micelle aggregate at 5 mg ml−1 increased from 366 for OSA-g-EPL8.5 to 1242 for OSA-g-EPL20.5. It suggested that the increase of substitution degrees caused more micelles to aggregate and form larger particles, which can also be verified by the PDF curves' peak shift towards a large value when the degree of substitution was increased. This result was also consistent with the relatively large particle sizes determined by dynamic light scattering.8
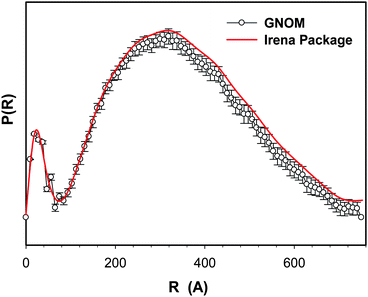 | ||
| Fig. 3 Comparison of pair distribution functions of OSA-g-EPL with 12.4% degree of substitution (DS) generated from either GNOM (empty circles) or Irena package (solid line). | ||
 | ||
| Fig. 4 Pair distribution function (PDF) curves of modified ε-polylysine with different degrees of substitution (DS): 8.5% (solid line); 12.4% (long dash line); and 20.5% (short dash line). | ||
| Dmax (Å) | Rg (Å) | Peak 1 (Å) | Peak 2 (Å) | naggregation | |
|---|---|---|---|---|---|
| OSA-g-EPL8.5 | 778 | 276 | 24 | 326 | 366 |
| OSA-g-EPL12.4 | 752 | 268 | 24 | 311 | 529 |
| OSA-g-EPL20.5 | 800 | 308 | 26 | 357 | 1242 |
Encapsulation of curcuminoids using M-EPL micelles
The hydrophobic cores of M-EPLs micelles may serve as a microenvironment to solubilize water-insoluble bioactive compounds and thus increase their water solubility. In the following section, curcuminoids were used as examples to illustrate the capability of M-EPL micelles to encapsulate and solubilize water-insoluble bioactive compounds.Loading water-insoluble compounds into the hydrophobic core of polymer micelles is usually controlled by kinetics. Different loading methods usually have different efficiencies.17 In this study, three loading methods (solvent evaporation, dialysis and homogenization) were compared to find the best way to achieve the highest loading capacity (and thus the water solubility) of curcuminoids in M-EPL micelles (Fig. 5). In the solvent evaporation method (Fig. 5A), chloroform with dissolved curcuminoids was homogenized to form a coarse emulsion in the M-EPL solution. As chloroform evaporated, a fraction of curcuminoids was trapped in the M-EPL micelles. In the dialysis method (Fig. 5B), M-EPL and curcuminoids were both dissolved in DMSO and dialyzed against dH2O. As the DMSO inside the dialysis bag was replaced by dH2O, curcuminoids were gradually encapsulated in M-EPL micelles. In the homogenization method (Fig. 5C), a simple high-speed homogenization was used. Under high shear, curcuminoids crystals were expected to be broken into smaller size, which, according to Ostwald-Freundlich equation, would have greater water solubility and thus be easier to be encapsulated into micelles. In our previous study, similar method was used to encapsulate curcumin into micelles formed by hydrophobically modified starch.12
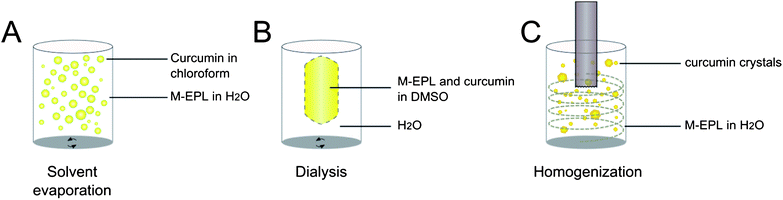 | ||
| Fig. 5 Scheme of three loading methods used to encapsulate curcuminoids into M-EPL micelles: (A) Solvent evaporation – curcuminoids were dissolved in chloroform and M-EPL was dissolved in distilled water (dH2O). Coarse emulsion was generated by high-speed homogenization. Subsequently, chloroform was removed by evaporation; (B) dialysis – curcuminoids and M-EPL were co-dissolved in DMSO and dialyzed again dH2O to remove DMSO; and (C) high-speed homogenization (HSH) – HSH was used to break down the curcuminoids crystals, and the high shear force facilitated curcuminoids dissolution and encapsulation. | ||
The encapsulated curcuminoids in the three methods were then quantified by HPLC. As clearly showed in Fig. 6, the dialysis method resulted in the highest loading capacity: in dried samples, curcuminoids accounted for 5.3 ± 1.9% (w/w), which was significantly higher than that in homogenization (1.1 ± 0.3%) and solvent evaporation methods (0.8 ± 0.4%). Accordingly, in 1 mg mL−1M-EPL micelle solution, the maximal solubilized curcuminoids concentration was 53 ± 19 μg/mL. Compared with the water solubility of curcuminoids (11 ng mL−1),18 this represented almost 5000-fold increase.
 | ||
| Fig. 6 Mass percentages of curcuminoids in freeze-dried M-EPL samples prepared by three loading methods described in Fig. 5. Data are presented as mean ± standard deviation (n = 3). * denotes statistically significant difference (P < 0.05). | ||
The percentages of the three curcuminoid compounds encapsulated in the micelles were also compared with that of curcuminoids raw materials (Table 2). It was found that the composition of curcuminoids prepared by dialysis was similar to that of original curcuminoids powder, while the composition of curcuminoids prepared by either solvent evaporation or high-speed homogenization had a slightly lower content of curcumin, suggesting that different loading methods may have weak preference toward selected curcuminoid components.
| Curcuminoids compound | Raw materials | Dialysis | High-speed homogenization | Solvent evaporation |
|---|---|---|---|---|
| a Data are presented as mean ± standard deviation (n = 3, except for raw materials, n = 12). | ||||
| Cur (%) | 82.1 ± 1.0 | 82.4 ± 1.3 | 77.7 ± 1.0 | 78.3 ± 5.4 |
| D-Cur (%) | 14.8 ± 0.5 | 14.8 ± 0.9 | 15.8 ± 0.4 | 13.9 ± 1.1 |
| BD-Cur (%) | 3.1 ± 0.5 | 2.8 ± 0.3 | 6.5 ± 0.6 | 7.8 ± 6.5 |
Additionally, dynamic light scattering was used to examine the particle size of the M-EPL micelles before and after encapsulation of curcuminoids: the hydrodynamic diameter of pure M-EPL micelle was 74.7 ± 1.0 nm with polydispersity of 0.424, while that of M-EPL micelle with curcuminoids was 135.5 ± 1.5 nm with polydispersity as 0.273. The results suggested that the encapsulation of curcuminoids can cause M-EPL micelles to associate and form larger micellar aggregates.
Differential scanning calorimetry (DSC) analysis
After micelle encapsulation, curcuminoids were thought to be solubilized in the core of the M-EPL micelles instead of existing as large crystals. To confirm the encapsulation and solubilization, DSC analyses were performed to detect curcuminoid crystals in the samples of curcuminoids powder, curcuminoids/M-EPL mixture, and curcuminoids in M-EPL micelles. As shown in Fig. 7A, curcuminoids powder was in crystal form and had a melting peak at 178 °C. Our HPLC analysis indicated that the curcuminoids powder contained about 82% curcumin, 15% DCur and 3% BDCur (w/w, Table 2). The melting points for curcumin, DCur and BDCur were reported as 184, 172 and 222 °C, respectively.19 Therefore, the melting peak of the curcuminoids powder at 178 °C may mainly arise from the compounded effect of curcumin and DCur. In the curcumin M-EPL mixture sample, much smaller melting peaks were also noticeable, as indicated by the arrow in Fig. 7A and more apparently in Fig. 7B, which showed two small peaks locating at 178 °C and 182 °C respectively. In contrast, there was no melting peak detected in the curcuminoids/M-EPL micelle sample, suggesting that curcuminoids were indeed encapsulated and solubilized in the M-EPL polymer micelle. | ||
| Fig. 7 (A) Differential scanning calorimetry (DSC) results of curcuminoids, curcuminoids/M-EPL simple mixture, and curcuminoids encapsulated in M-EPL micelle prepared by dialysis method. The arrow indicates the melting peaks of curcuminoids in the mixture; and (B) Zoom-in DSC curves of simple mixture of curcuminoids and M-EPL as well as curcuminoids encapsulated in M-EPL micelle through dialysis in the temperature from 160 to 200 °C. | ||
The DSC results also suggest that compared with other types of formulations, such as solid lipid nanoparticle and liposome, this micelle encapsulation system is ready to be lyophilized and reconstituted.
M-EPL micelle encapsulation stabilized curcuminoids at pH 7.4
It is well known that although curcumin has higher water solubility in basic aqueous solutions, it undergoes rapid hydrolysis.20–23 In this study, we examined the effect of M-EPL encapsulation on preventing curcuminoids hydrolysis at pH 7.4. As shown in Fig. 8A, the result suggested that free curcuminoids underwent rapid hydrolysis, while encapsulation in M-EPL micelles was able to stabilize curcuminoids as other micelle systems formed by small-molecular-weight surfactants.20,21 | ||
| Fig. 8 Stability of curcuminoids at pH 7.4: (A) The stability of total curcuminoids for the free curcuminoids versus curcuminoids encapsulated in M-EPL micelle through dialysis; and (B) The stability of curcumin, demethoxycurcumin (DCur), and bisdemethoxycurcumin (BDCur) for their free forms versus each component encapsulated in M-EPL micelle through dialysis. Data are shown as mean ± standard deviation (n = 3). | ||
Since the curcuminoids used in this study contained curcumin, DCur and BDCur, the stability of each individual curcuminoid compound with and without encapsulation was examined simultaneously. As shown in Fig. 8B, different curcuminoid compounds had different hydrolysis rate at pH 7.4. Namely, curcumin underwent fastest hydrolysis, followed by DCur, while BDCur was relatively resistant to the hydrolysis. To the best of our best knowledge, this was the first time that different curcuminoids showed different stability against hydrolysis at weak alkaline conditions. Since BDCur was relatively stable and soluble at pH 7.4, its bioactivity and bioavailability compared with that of curcumin may need further examination.
Encapsulated curcuminoids showed elevated cellular antioxidant activity
Curcuminoids, especially curcumin, with their phenolic groups, beta-diketone and double bonds structure, are well-known antioxidant compounds. Using a recently-developed cell-based method,24 the cellular antioxidant activity (CAA) of curcuminoids was measured. As shown in Fig. 9, the EC50 value of curcuminoids was determined as 4.4 μM, which was among the most potent natural antioxidants.24 Furthermore, the CAA values of encapsulated curcuminoids were compared with that of free curcuminoids at the concentration of 2 μM, at which, the CAA value of M-EPL used for encapsulation was close to zero (Fig. 10). It was clearly shown that encapsulated curcuminoids had greater CAA value than free curcuminoids, suggesting M-EPL micelle encapsulation may serve as a good delivery system for curcumin(oids) and other water-insoluble bioactives. Although it would be ideal to determine the EC50 of encapsulated curcuminoids, M-EPL at higher concentrations exhibited various antioxidant activities (not shown) and may interfere with the interpretation of the CAA results for the encapsulated curcuminoids. Therefore, only the situation where modified EPL has negligible CAA value was shown and the CAA values of free curcuminoids and encapsulated curcuminoids were compared. | ||
| Fig. 9 Measurement of the cellular antioxidant activity of curcuminoids: (A) Cellular antioxidant activity (CAA) of curcuminoids at different concentrations; and (B) Determination of the EC50 of curcuminoids. Data are shown as mean ± standard deviation (n = 3). | ||
 | ||
| Fig. 10 Comparison of the CAA values of free curcuminoids and encapsulated curcuminoids. CAA values were determined at the curcuminoids concentration of 2 μM. Data are presented as mean ± standard deviation (n = 4). ** denoted for very significant difference (p < 0.001, t-test). | ||
Two mechanisms may be used to explain the enhanced cellular antioxidant activity upon encapsulation. First, free curcuminoids were technically dispersed from DMSO solution into the cell media, which may form sub-micron sized particles and have limited solubility. Encapsulated curcuminoids, on the other hand, were originally in the dissolved form in the micelle core and may still largely remain soluble form upon dilution in the treatment media. Therefore, the concentration of dissolved curcuminoids was expected to be greater in curcuminoids micelle solution than in curcuminoids dispersion. The second possible mechanism may be the rapid hydrolysis of curcuminoids at weak basic condition. Treatment media were at pH 7.4, which may cause rapid degradation of curcuminoids, as shown in Fig. 8. On the other hand, encapsulation may stabilize curcuminoids against hydrolysis. Thus, the curcuminoids amount and the cellular antioxidant activity from the micelle encapsulation were expected to be greater. On the other hand, these interpretations did not exclude the possibility that M-EPL had specific interaction with the HepG2 cells which might facilitate the movement of curcuminoids into/onto the cells.
Conclusion
In summary, modified ε-polylysine was able to form polymer micelles. From the SAXS analysis, the M-EPL micelles were spherical and able to further form micelle aggregates. This micelle system was used to encapsulate curcuminoids. It was demonstrated that dialysis method generated a higher loading capacity than either the solvent evaporation or high-speed homogenization method. The micelle encapsulation was confirmed as no crystals of curcuminoids were detected after encapsulation. Meanwhile, it was shown that upon encapsulation, curcuminoids were stabilized against hydrolysis at pH 7.4, and had enhanced cellular antioxidant activity compared with free curcumin. This work suggested that M-EPL may serve as a delivery system for curcuminoids and other water-insoluble nutraceuticals.Acknowledgements
This work is supported by United States Department of Agriculture – Agriculture and Food Research Initiative (USDA-AFRI) grant (2009-65503-05793), and in part by Advanced Orthomolecular Research, Inc. (AOR).References
- G. S. Kwon and T. Okano, Adv. Drug Delivery Rev., 1996, 21, 107–116 CrossRef CAS.
- V. P. Torchilin, J. Controlled Release, 2001, 73, 137–172 CrossRef CAS.
- B. B. Aggarwal, A. Kumar and A. C. Bharti, Anticancer Res., 2003, 23, 363–398 CAS.
- B. Joe, M. Vijaykumar and B. R. Lokesh, Crit. Rev. Food Sci. Nutr., 2004, 44, 97–111 CrossRef CAS.
- A. Duvoix, R. Blasius, S. Delhalle, M. Schnekenburger, F. Morceau, E. Henry, M. Dicato and M. Diederich, Cancer Lett., 2005, 223, 181–190 CrossRef CAS.
- R. A. Sharma, A. J. Gescher and W. P. Steward, Eur. J. Cancer, 2005, 41, 1955–1968 CrossRef CAS.
- P. Anand, A. B. Kunnumakkara, R. A. Newman and B. B. Aggarwal, Mol. Pharmaceutics, 2007, 4, 807–818 CrossRef CAS.
- H. Yu, Y. Huang and Q. Huang, J. Agric. Food Chem., 2010, 58, 1290–1295 Search PubMed.
- I. L. Shih, M. H. Shen and Y. T. Van, Bioresour. Technol., 2006, 97, 1148–1159 Search PubMed.
- M. Saimura, M. Takehara, S. Mizukami, K. Kataoka and H. Hirohara, Biotechnol. Lett., 2008, 30, 377–385 Search PubMed.
- USFDA, in GRAS Notice No. GRN 000135, ed. U. S. F. a. D. Administration, Washington, D.C., 2004 Search PubMed.
- H. Yu and Q. Huang, Food Chem., 2010, 119, 669–674 Search PubMed.
- K. L. Wolfe, X. M. Kang, X. J. He, M. Dong, Q. Y. Zhang and R. H. Liu, J. Agric. Food Chem., 2008, 56, 8418–8426 Search PubMed.
- J. Ilavsky and P. R. Jemian, J. Appl. Crystallogr., 2009, 42, 347–353 CrossRef CAS.
- D. Svergun, J. Appl. Crystallogr., 1992, 25, 495–503 CrossRef.
- Y. Huang, H. Yu, L. Guo and Q. Huang, J. Phys. Chem. B, 2010, 114, 7719–7726 Search PubMed.
- M.-C. Jones and J.-C. Leroux, Eur. J. Pharm. Biopharm., 1999, 48, 101–111 CrossRef CAS.
- Y. Kaminaga, A. Nagatsu, T. Akiyama, N. Sugimoto, T. Yamazaki, T. Maitani and H. Mizukami, FEBS Lett., 2003, 555, 311–316 Search PubMed.
- L. Peret-Almeida, A. P. F. Cherubino, R. J. Alves, L. Dufoss and M. B. A. Gloria, Food Res. Int., 2005, 38, 1039–1044 Search PubMed.
- H. H. Tonnesen, Pharmazie, 2002, 57, 820–824 CAS.
- M. H. M. Leung, H. Colangelo and T. W. Kee, Langmuir, 2008, 24, 5672–5675 Search PubMed.
- Z. Wang, M. H. M. Leung, T. W. Kee and D. S. English, Langmuir, 2009, 26, 5520–5526 Search PubMed.
- M. H. M. Leung and T. W. Kee, Langmuir, 2009, 25, 5773–5777 Search PubMed.
- K. L. Wolfe and R. H. Liu, J. Agric. Food Chem., 2007, 55, 8896–8907 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2011 |
