A generalized probe selection method for DNA chips†
Satish Balasaheb
Nimse
a,
Keum-Soo
Song
b,
Junghoon
Kim
b,
Van-Thao
Ta
a,
Van-Thuan
Nguyen
a and
Taisun
Kim
*a
aInstitute for Applied Chemistry and Department of Chemistry, Hallym University, Chuncheon, 200-702, Korea. E-mail: tskim@hallym.ac.kr; Fax: +82-33-256-3421
bBiometrix Technology, Inc., 202 BioVenture Plaza, Chuncheon, 200-161, Korea
First published on 6th October 2011
Abstract
The flaws in the present probe selection methods restrained the development of the DNA chip technology and its applications. The presented generalized probe selection method for the DNA chips elaborates the length of the probe, the melting temperatures, the specificity of the probe, and the position where the probe may bind to the targets.
The development of DNA chips in the last decade has not only revolutionized the fields of genetics and genomics but also the field of medical diagnostics in terms of identification and discrimination of the pathogens in infectious diseases.1 The selected probes immobilized on the solid support produce a variety of DNA chips.2 A computational approach for the probe selection has been studied extensively.3 But this approach suggests at least 6–10 different probes with varying lengths for the single genotype,4 which results in the delay of the technological advancement at the expense of the valuable resources.5
However, there is no report on a generalized probe selection method, which may include the length of the probe, the specificity of the probe, and the position where the probe may bind to the targets.6 Based on the literature it is noticed that the probe sequence is chosen in such a way that it binds in the middle of the PCR region of the target DNAs.7 The sensitivity and the specificity of the DNA chip depend on the favorable hydrogen bonding between the complementary base pairs in the probe and target oligonucleotide sequences during hybridization.8 The consensus of the hydrogen bonding is that it is stronger at a low temperature as compared to elevated temperatures. Reported methods suggest the elevated temperatures of 40–60 °C with the longer hybridization time ranging from a few hours to overnight hybridization.9 This is probably due to the compromise which has to be made to obtain the good specificity at the expense of the sensitivity.10
Therefore, to boost the research based on the DNA chip technology, the generalized probe selection method is highly indispensable. Based on our study on the 9G DNAChips,11 the hybridization pattern (Scheme 1) of the immobilized probes with the target and non-target Cy5 labeled DNAs shows that at least two mismatches one in the top and one in the bottom of the immobilized probe are required to remove the non-target hybridizations.
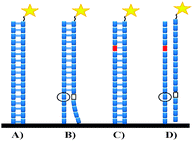 | ||
| Scheme 1 Hybridizations of the immobilized probes with the Cy5 labeled target DNAs. (A) Original probe–target specific hybridization, (B) original probe–target non-target hybridization, (C) original probe (with one artificial mutation)-target specific hybridization, (D) original probe (with one artificial mutation)–target no non-target hybridization. | ||
Here, we introduce a generalized probe selection method for DNA chips, which involves the use of single artificial mutation in the immobilized probes. The presented probe selection method is explained by establishing the proficient genotyping of the 19 different types of the human papillomavirus (HPV). Moreover, the application of this method is confirmed by analyzing more than 1000 clinical samples.
According to our previous report on the 9G DNAChips, different types of 9G DNAChips were produced by immobilizing Probe1–Probe82 (see the ESI†, Table S1) on the aminocalix[4]arene (AMCA) slide. To find the optimum length of the probe to be immobilized on the chip surface, the probes (Probe1–Probe6) complementary to the sequences of HPV16 and HPV18 with the length of the 14, 17, and 20 mers were immobilized and hybridized with 100 fmol of the Cy5 labeled PCR products of HPV16 and HPV18 (see the ESI†, Fig. S6).
The probes with 17 mers and 20 mers showed a strong fluorescence intensity after hybridization with the Cy5 labeled PCR products as compared to the probes with 14 mers (see the ESI†, Fig. S2, S3, and S6). The use of probes with 17 mers for the fabrication of the 9G DNAChip is more rational as it can avoid the non-target hybridization, which may arise due to three extra nucleotides in the probes with the 20 mers. However, to compensate for keeping the melting temperature in the range of 45 to 51 °C the probes with the 20 mer sequence can be used. The probe length being selected to 17 mer, it was important to select the position on the genome to start counting for the 17 mers.
The MY11/GP6+ primer set consists of a fixed nucleotide sequence for the forward primer and reverse primer, respectively, which amplifies 189 bp DNA fragments in the conserved L1 region (Fig. S1, ESI†) and detects a wide range of HPV types by using a lowered annealing temperature during PCR.12 Therefore, we focused on the HPV genome oligomer sequence near the region of the complementary sequence of the reverse primer for the selection of the probes for MY11/GP6+(M2) primer set-mediated PCR products.
The first nucleotide base on the genome complementary to the last nucleotide base in the reverse primer is considered as “0”. Each consecutive nucleotide base on the right and left of the “0” is numbered as −1 to −14 and 1 to 11, respectively (Fig. 1).
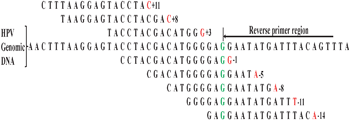 | ||
| Fig. 1 Selection of the probes complementary to the genomic DNA sequence. | ||
Hence, the sequence containing one base pair overlaps with the primer region became −1Probe, with five base pair overlaps −5Probe, similarly we chose −8Probe, −11Probe, and −14Probe. The first nucleotide base after ‘0’ on the left become +1Probe, after three nucleotide bases +3Probe, similarly we chose +5Probe, +8Probe, and +11Probe. Following this method we chose the probes −14 (Probe7, Probe15), −11 (Probe8, Probe16), −8 (Probe9, Probe17), −5 (Probe10, Probe18), −1 (Probe11, Probe19), +3 (Probe12), +5 (Probe20), +8 (Probe13, Probe21) and +12 (Probe14, Probe22) for the HPV genotype 16 (HPV16) and HPV genotype 18 (HPV18).
The selected probes (Probe7–Probe22) with a length of the 17 mers (Fig. 1) starting from the −14, −11, −8, −5, −1, +3, +7, and +11 positions on the genome of HPV16 and HPV18 were immobilized on the AMCA slide, and hybridized with 100 fmol of the Cy5 labeled PCR products of HPV16 and HPV18 (see the ESI†, Fig. S4–S6). The signal intensities shown by both −1 and +3 probes after hybridization with the Cy5 labeled PCR products of HPV16 and HPV18 are similar. Therefore it was considered that the Probes with 17 mer starting from the −1, or +3 positions can be used for the construction of the microarray.
It is interesting to notice that the probe starting from the −1 position has a G (guanine) subunit in all 19 genotypes of HPV. However, in the case of +3 or +5 positions, only few of the genotypes have G. It is well known that the guanine shows good hydrogen bonding and contributes more to the melting temperature (TM) of the oligomers. Therefore, we decided to go with the −1 position and hence 19 different probes (Probe23–Probe41) complementary to the 19 HPV genotypes were selected and used to produce Pre-HPV 9G DNAChips (Fig. 2B). The TM of the probes (Probe23–Probe41) is in the range of 45 to 51 °C.
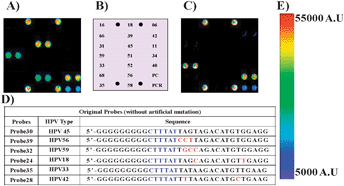 | ||
| Fig. 2 (A) Fluorescence image after the hybridization of the immobilized probes (Probe23–Probe41) with the Cy5 labeled PCR product of HPV45, (B) depicts the scheme for the immobilization of the probes (Probe23–Probe41) to obtain Pre-HPV 9G DNAChips. (C) Fluorescence image after the hybridization of immobilized probes (Probe23–Probe41) with the Cy5 labeled PCR product of HPV33, (D) a table depicting the sequences of the probes, (E) fluorescence scale, PMT gain = 48. | ||
The hybridization of 100 fmol of the Cy5 labeled PCR products of all 19 HPV genotypes with the probes on the Pre-HPV 9G DNAChips not only showed a good hybridization ability with the respective probes but also showed the non-target hybridizations with other probes (Fig. 2A). However, Probe35 (HPV33), Probe37 (HPV40), and Probe29 (HPV39) did not show any non-target hybridizations (Fig. 2C, and see the ESI†, Fig. S7–S25).
When the Pre-HPV 9G DNAChip was hybridized with the Cy5 labeled PCR product of HPV45 the spots corresponding to HPV56 and HPV59 were also appeared but with the lower fluorescence intensities as compared to the spots corresponding to HPV45 (Fig. 2A and E). By close observation of the sequences of Probe30 (HPV45), Probe39 (HPV56), and Probe32 (HPV59) with the TM ranging from 45 to 49 °C, it can be noticed that Probe30, Probe39, and Probe32 have top 13 to 14 similar nucleotides from the 3′-end in their respective sequences and at least three different nucleotides from 14 to 17 (Fig. 2D). Therefore, it is obvious that the similarity between the probe sequences resulted in the non-target hybridizations. However, the small difference in probe sequence significantly lowers the TM of Probe39 and Probe32 as compared to Probe30, eventually reducing the hybridization efficiency of these probes towards the PCR product of HPV45.
Probe24 (HPV18) has only two different nucleotides in its sequence with respect to Probe30 (HPV45), one in the top nine nucleotides and one in the bottom 13 to 17 nucleotides from the 3′-end. It is interesting to notice that Probe24 did not show any non-target hybridization with the Cy5 labeled PCR product of HPV45. A similar pattern was also found in the case of Probe35 (HPV33) and Probe28 (HPV42). The reason behind this could be the two base pair mismatches one in the top and the other in the bottom sequence which leads to the significant decrease in the TM, eventually resulting in the very poor hybridization efficiency of Probe24 and Probe28 towards the Cy5 labeled PCR products of HPV45 and HPV33, respectively.
The probes which showed non-target hybridizations had similarity between the top nine nucleotides from the 3′-end in their respective sequences. However, the probes which did not show any non-target hybridizations had at least one or more base-pair (bp) mismatches in their top nine nucleotides as well as in the bottom nine nucleotides. The trend of this specific and non-target hybridizations among the immobilized probes has laid down a simple rule, when there is at least one mismatch in the top nine and another one in the bottom nine no significant non-target hybridization will be observed. Therefore, to confront this rule, we decided to put one artificial mutation in the probes.
The nucleotide T (Thymine) was used to replace the original nucleotides in the positions 1–9 of Probe23 complementary to HPV16 to get the probes with one (Probe42–Probe50), two (Probe51–Probe56) and three (Probe57–Probe59) mutations. Probe23, and Probes42–Probe59 were immobilized on the AMCA slide and hybridized with 100 fmol of the Cy5 labeled PCR product of HPV16 (see the ESI†, Fig. S26). The probes with two mismatches (Probe51–Probe56) showed almost eight times decreased fluorescence intensity as compared to the original probe (Probe23). Whereas, the probes containing three mismatches (Probe57–Probe59) did not show any hybridization. It is interesting to notice that the probes with one mutation at different positions showed drastic changes in the hybridization efficiency with the Cy5 labeled PCR product of HPV16. The probes with mutations at 3 to 7 positions showed approximately five times decrease in the fluorescence intensity as compared to the original probe (Probe23). These results clearly indicate that to reduce or eliminate the non-target hybridizations it is necessary to put at least one artificial mutation at positions 3 to 7.
To demonstrate the applicability of the presented probe selection method, the 19 different probes (Probe60–Probe78) with one artificial mutation at positions 3 to 7 were immobilized to obtain the HPV 9G DNAChips and hybridized with Cy5 labeled PCR products of all 19 HPV genotypes (Fig. 3 and also see the ESI†, Fig. S27–S34).
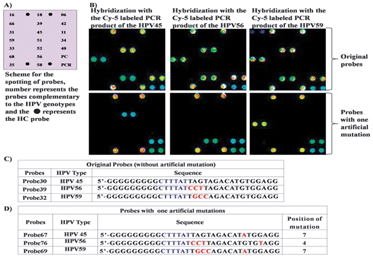 | ||
| Fig. 3 (A) Depicts the scheme for the immobilization of the probes (Probe60–Probe78) to obtain HPV 9G DNAChips, (B) fluorescence image after the hybridization with the Cy5 labeled PCR product of HPV45, HPV56, and HPV59 (top: probes without mutation, bottom: probes with one artificial mutation), (C) and (D) depict the sequences of the probes without artificial mutation and with artificial mutation, respectively, PMT gain = 48. | ||
After hybridization of the HPV 9G DNAChip with the Cy5 labeled PCR products of HPV45, HPV56, and HPV59 the non-target hybridization with other HPV types (Fig. 3B, fluorescence images at the bottom) was absent. The use of single artificial mutation in Probe30, Probe39, and Probe32 with the TM of 45, 47, and 49 °C, respectively, clearly demonstrates the increase in the specificity of the selected probes (see the ESI†, Fig. S35). The probes with TM 45 °C showed 40% decreased intensity and the probe with TM 47 °C showed 10% decreased intensity. The probes with TM 49 °C showed no significant intensity change. The probes (Probe60–Probe78) with single artificial mutation showed 200 times stronger signal intensity for the target hybridization as compared to the non-target hybridization (see the ESI†, Fig. S35).
The HPV 9G DNAChips were employed for the identification and discrimination of the 19 different HPV genotypes in the clinical samples. The results of the clinical study established the performance characteristic of HPV 9G DNAChips for the HPV test (see the ESI† Table S2 and S3). The genotyping results by HPV 9G DNAChips were 100% identical with the sequencing data of the same samples.
In this article, we have presented a generalized probe selection method for the 9G DNAChips, which includes the length of the probe (17 mers to 20 mers), the melting temperatures (47 to 51 °C), the specificity of the probe (it is necessary to put at least two mutations one in the top nine and the other in the bottom nine), and the position where the probe may bind to the targets (at least one overlap with the primer region). The excellent specificity of the probes designed by the presented probe selection method is ensured by the use of the single artificial mutation in the immobilized probes which provide 200 times stronger signal intensity for the target hybridization as compared to the non-target hybridization.
The presented probe selection method not only accelerates the technological advancement for the high-performance DNA chips but also enables the research scientists to efficiently use the valuable resources.
This work was supported by the Ministry of Knowledge and Economy. T. Kim acknowledges Dr H. Ahn of the CHA clinic for providing the clinical samples.
Notes and references
- E. Hodges, Z. Xuan, V. Balija, M. Kramer, M. N. Molla, S. W. Smith, C. M. Middle, M. J. Rodesch, T. J. Albert, G. J. Hannon and R. W. McCombie, Nat. Genet., 2007, 39, 1522 CrossRef CAS.
- A. C. Pease, D. Solas, E. J. Sullivan, M. T. Cronin, C. P. Holmes and S. P. Fodor, Proc. Natl. Acad. Sci. U. S. A., 1994, 91, 5022 CrossRef CAS.
- S. B. Roth, J. Jalava, O. Ruuskanen, A. Ruohola and S. Nikkari, J. Clin. Microbiol., 2004, 42, 4268 CrossRef CAS.
- M. A. Reyes-Lopez, A. Mendez-Tenorio, R. Maldonado-Rodriguez, M. J. Doktycz, J. T. Fleming and K. L. Beattie, Nucleic Acids Res., 2003, 31, 779 CrossRef CAS.
- Y. Huang, C. Chang, C. Chan, T. Yeh, Y. Chang, C. Chen and C. Kao, Bioinformatics, 2005, 21, 4330 CrossRef CAS.
- R. F. Wang, M. L. Beggs, L. H. Robertson and C. E. Cerniglia, FEMS Microbiol. Lett., 2002, 213, 175 CrossRef CAS.
- R. Peytavi, L. Tang, F. R. Raymond, K. Boissinot, L. Bissonnette, M. Boissinot, F. J. Picard, A. Huletsky, M. Ouellette and M. G. Bergeron, BioTechniques, 2005, 39, 89 CrossRef CAS.
- N. L. W. Franssen-van Hal, O. Vorst, E. Kramer, R. D. Hall and J. Keijer, Anal. Biochem., 2002, 308, 5 CrossRef CAS.
- A. J. Dingley, R. D. Peterson, S. Grzesiek and J. Feigon, J. Am. Chem. Soc., 2005, 127, 14466 CrossRef CAS. ***
- O. V. Matveeva, S. A. Shabalina, V. A. Nemtsov, A. D. Tsodikov, R. F. Gesteland and J. F. Atkins, Nucleic Acids Res., 2003, 31, 4211 CrossRef CAS.
- (a) K. Song, S. B. Nimse, J. Kim, J. Kim, V. Ta, V. Nguyen and T. Kim, Chem. Commun., 2011, 47, 7101 RSC; (b) K. Song, S. B. Nimse, J. Kim, J. Kim, V. Ta, V. Nguyen and T. Kim, Chem. Commun., 2011, 47, 7716 RSC.
- A. L. F. Haws, Q. He, P. L. Rady, L. Zhang, J. Grady, T. K. Hughes, K. Stisser, R. Konig and S. K. Tyring, J. Virol. Methods, 2004, 122, 87 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available: Materials and method, Table S1–S3, Fig. S1–S34, detection and discrimination HPV genotypes in clinical samples. See DOI: 10.1039/c1cc15137a |
| This journal is © The Royal Society of Chemistry 2011 |
