 Open Access Article
Open Access Article9G DNAChip: a platform for the efficient detection of proteins†
Keum-Soo
Song
a,
Satish Balasaheb
Nimse
b,
Junbae
Kim
b,
Junghoon
Kim
a,
Van-Thao
Ta
b,
Van-Thuan
Nguyen
b and
Taisun
Kim
*b
aBiometrix Technology Inc., 202 BioVenture Plaza, Chuncheon, 200-161, Korea
bInstitute for Applied Chemistry and Department of Chemistry, Hallym University, Chuncheon, 200-702, Korea. E-mail: tskim@hallym.ac.kr; Fax: +82-33-256-3421
First published on 8th June 2011
Abstract
According to the proposed DAGON method, the CRP and PSA antigens with the concentrations of 1 pg ml−1 to 10 pg ml−1 range can be easily differentiated in the buffer matrix. Moreover, it is for the first time that the multiple antigens with the concentrations of 1 pg ml−1 and 0.1 pg ml−1 can be detected in the mixture of the proteins without an amplification technique.
Cancer is the leading cause of morbidity and mortality in the United States and Europe.1 Cancer can be diagnosed at an early stage with the help of efficient biomarker detection techniques.2Protein microarrays have been developed for the detection of the biomarkers.3
The protein microarrays produced by the direct immobilization methods are known to suffer from drawbacks like the instability of the immobilized proteins, thus resulting in low sensitivity.4,5 DNA-Directed Immobilization (DDI) was employed to improve the stability of the proteins by immobilizing them on the surface shortly before the detection of antigens.6 In such stepwise methods, first the proteins are immobilized on the surface and in the second step they are allowed to react with the target proteins. The disadvantage of this method is: once the proteins are immobilized on the surface they still have a chance to lose their activity over the period of time.7 Thus, methods like DDI improve the stability of the proteins, but the lengthy process limits the sensitivity of the method to 100 pg ml−1.8
To improve the sensitivity of the detection method, many signal amplification techniques such as the bio-barcode and immuno-PCR have been used, albeit each has its own advantages and drawbacks.9 In our previous report, we have demonstrated that the conventional immobilization methods can also result in the loss of the activity of the analyte proteins.10 Hence, we propose that the problems related to the direct immobilization of the proteins can be minimized by allowing the antigen, antibody biomolecular complexes to be formed in the solution and hybridize to the specific probes on the 9G DNAChip surface for the detection. Here, we report on the efficient detection of the antigens like CRP and PSA. The normal levels of the CRP and PSA antigens are more than 1 ng ml−1 in the serum. Hence to detect the antigen concentrations below 100 to 1 pg ml−1, we used the buffer matrix resembling the serum.
Recently, we have reported the 9G DNAChip, which shows more than 80% hybridization efficiency at 25 °C in 5 min with specificity of more than 97%. 9G DNAChips were produced by the immobilization of Probe 1–Probe 11 (see the ESI†, Table S1 and Scheme S1) following the reported method.11 The IgG antibody (IgGAb), CRP antibody (CRPAb), and PSA antibody (PSAAb) were conjugated with the specific oligomers T1, T2, and T3, respectively (see the ESI†, Table S1). The secondary antibodies of CRP (CRPAB) and PSA (PSAAB) were labeled with Cy5 dyes. The CRP antigen (CRPAg) and PSA antigen (PSAAg) were used to probe the proposed method (see the ESI† for abbreviations).
Scheme 1 depicts the hybridization and detection of the CRP and PSA antigens by the DNA-Guided Detection method. The method can be explained in detail as follows. The biomolecular complex of the Cy5-labeled secondary antibody, the antibody–DNA conjugate and the target antigen formed in the solution is site-specifically guided to the predestined area on the chip surface and hybridized at the room temperature.
 | ||
| Scheme 1 DNA-guided detection (DAGON) method. | ||
First, the hybridization mixture was prepared by mixing the different Cy5-labeled antibodies (Cy5-CRPAB, Cy5-PSAAB) and antibody–DNA conjugates (IgG-T1, CRPAb-T2, and PSAAb-T3) in the hybridization solution. With or without the addition of the antigens to the hybridization mixture, the IgG-T1 and the labeled antibody (Cy5-CRPAB or Cy5-PSAAB) form a complex (Cy5-CRPAB-IgGT1 or Cy5-PSAAB-IgGT1) which is hybridized on the specific site after loading on the 9G DNAChip surface. Therefore, the IgG-T1 complexed with the Cy5-labeled antibodies (Cy5-CRPAB, Cy5-PSAAB) serves as an internal standard, as it is indicative of the formation of a biomolecular complex in a solution.
With the addition of the CRPAg and the PSAAg to the mixture of the Cy5-CRPAB, Cy5-PSAAB, IgG-T1, CRPAb-T2, and PSAAb-T3, they form respective complexes Cy5-CRPAB-CRPAg-CRPAb-T2, Cy5-PSAAB-PSAAg-PSAAb-T3 in the solution. After loading on the 9G DNAChip surface, the target DNA's T1, T2, and T3 guide the respective biomolecular complexes to the specific sites and they are hybridized with the surface-bound complementary oligomers for the detection.
The 50 μl of the hybridization mixture was prepared by mixing 5 μl each of the IgGAb-T1 (0.2 μg ml−1), CRPAb-T2 (25 μg ml−1), Cy5-CRPAB (25 μg ml−1), PSAAb-T3 (25 μg ml−1), Cy5-PSAAB (25 μg ml−1), and Cy5-T4 (40 fmol μl−1) with the appropriate volume of the hybridization solution (see the ESI†, Table S2). Individual antigen detections were carried out by mixing 5 μl (0.001 ng ml−1–10 ng ml−1) of the CRPAg or PSAAg in the 45 μl of the hybridization mixture to get final concentrations of the antigens to 0.1 pg ml−1–1 ng ml−1 and allowed to stand for 5 min. During this time, the biomolecules form their respective biomolecular complexes, such as Cy5-CRPAB-CRPAg-CRPAb-T2 and Cy5-PSAAB-PSAAg-PSAAb-T3 in the solution. Then the mixture was loaded in the hybridization chambers formed by covering the 9G DNAChips with Secure-Seal™, and then allowed to hybridize for 30 min at 25 °C. After washing and drying, the 9G DNAChips were scanned and analyzed. For the simultaneous detection, 5 μl (0.001 ng ml−1–10 ng ml−1) of each of the CRPAg and PSAAg were mixed with the 40 μl of the hybridization mixture and the same procedure was followed for the individual detection. The mixture with six different concentrations of the antigen was loaded on the single 9G DNAChip at the same time and processed. The obtained results are presented in Fig. 1.
 | ||
| Fig. 1 (A) Individual detections of CRPAg and PSAAg with the concentrations of 0.1 pg ml−1–1 ng ml−1, (B) a graph representing the fluorescence intensities for CRPAg and PSAAg at different concentrations for individual detection, (C) simultaneous detection of CRPAg and PSAAg with the concentrations of 0.1 pg ml−1–1 ng ml−1, (D) a graph representing the fluorescence intensities for CRPAg and PSAAg at different concentrations for simultaneous detection. PMT gain = 90. | ||
As shown in Fig. 1A and B, the biomolecular complex of IgG-T1 (Cy5-CRPAB-IgG-T1 or Cy5-PSAAB-IgG-T1) with the Cy5-CRPAB or Cy5-PSAAB is site-specifically guided and hybridized to the complementary Probe 1 on the chip surface. In a similar way, the biomolecular complexes Cy5-CRPAB-CRPAg-CRPAb-T2 and Cy5-PSAAB-PSAAg-PSAAb-T3 are site-specifically guided and hybridized to the complementary Probe 3 and Probe 5 on the chip surface, respectively (Fig. 1C, 3A).
The detection of the low-abundance proteins in the presence of the other proteins is an essential criterion for the diagnostic purposes. Hence, we performed the simultaneous detection of the CRPAg and the PSAAg. As shown in Fig. 1C and D, the biomolecular complexes Cy5-CRPAB-CRPAg-CRPAb-T2 and Cy5-PSAAB-PSAAg-PSAAb-T3 in the solution are site-specifically guided and hybridized to the respective positions on the chip surface. Fig. 1 clearly demonstrates that the DAGON method allows the individual or simultaneous detection of the antigens with a concentration as low as 1 pg ml−1.
The fluorescence intensities of the antigen concentrations (CRPAg, PSAAg) between 10 pg ml−1 to 1 pg ml−1 can be clearly distinguished at the PMT gain = 90. The fluorescence intensities of the antigen concentrations between 100 pg ml−1 to 10 pg ml−1 can also be clearly distinguished at the PMT gain = 70. Fig. 1C and 2 illustrate that the ratios of the fluorescence intensities of 100 pg ml−1 to 10 pg ml−1 and 10 pg ml−1 to 1 pg ml−1 are similar in both antigens.
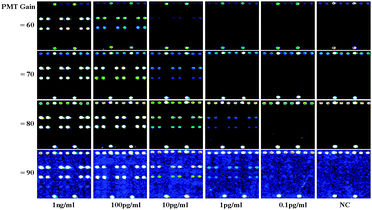 | ||
| Fig. 2 Fluorescence images for the detection of CRPAg (on the Lane 3) and PSAAg (on the Lane 5) with concentrations of 0.1 pg ml−1–1 ng ml−1 at different PMT gains. | ||
The detection limits of the protein microarrays are significantly limited due to the loss of the activities of the immobilized proteins.8 To investigate the applicability of the DAGON method for the simultaneous detection of the CRPAg and PSAAg and to find if there is any nonspecific interaction the 9G DNAChips were scanned at different PMT gains, the results are shown in Fig. 2.
Fig. 2 demonstrates that by increasing the PMT gains from 60 to 90 it is possible to detect the 1000 times lower concentrations of the antigens. At the PMT gain 90, the antigen with 0.1 pg ml−1 concentration can also be detected. The results are comparable to the negative control (NC, without addition of any antigens). Fig. 2 also reveals very high specificity, as the Cy5-CRPAB-CRPAg-CRPAb-T2 and Cy5-PSAAB-PSAAg-PSAAb-T3 were site-specifically guided to Probe 3 and Probe 5 on the 9G DNAChip, respectively.
The detection limits of the protein microarrays are significantly limited due to the signals for the non-specific interactions.12 If the false signals for the non-specific interactions are minimized, any signals higher than that of the NC can be used for the effective detection of the antigens. Therefore, it is very important to be sure that there are no non-specific interactions in the detection of the antigens at the sub-picogram level.
Fig. 3B, C and D demonstrate the fluorescence maps for the detection of the PSAAg at 1 ng ml−1, 10 pg ml−1 and NC (without PSAAg), respectively. The fluorescence intensities of the spotted area of NC and the background (non-spotted area) in each case are around 7000–6400 and 6700–6200, respectively. Hence, the similar fluorescence intensities for the spotted areas and the background indicate that there are no non-specific interactions. It is also important to notice that the fluorescence intensity of the PSAAg at 0.1 pg ml−1 is in the range of 7800–8200.
 | ||
| Fig. 3 (A) Schematic of the 9G DNAChip with immobilized probes (Probe 1–Probe 11), Probe 5 complementary to the target biomolecular complex Cy5-PSAAB-PSAAg-PSAAb-T3, (B) fluorescence image for the detection of PSAAg (1 ng ml−1), (C) fluorescence image for the detection of PSAAg (10 pg ml−1), (D) fluorescence image of the negative control in the detection of PSAAg, PMT gain = 90. | ||
The presented method shows the efficient detection of 0.1 pg ml−1 of the PSA and CRP antigens with 100% specificity. There were no measurable hybridization signals from the spots of the noncomplementry (Probe 2) probe, indicating that the nonspecific hybridization did not occur. The major reasons for such a high sensitivity and specificity could be the formation of the biomolecular complex in the solution, and the hybridization, washing, and drying at room temperature. Furthermore, the ultrafast hybridization (80% in 30 min at 25 °C) of target probes in the biomolecular complexes with the specific probes on the 9G DNAChip at room temperature reduces the chances of the non-specific interactions (see the ESI†, Fig. S1).
In this communication, we have demonstrated the method for the efficient detection of the antigens. According to the DAGON method, the antigens with the concentrations of 1 pg ml−1 to 10 pg ml−1 range can be easily differentiated. Moreover, it is for the first time that the multiple antigens with the concentrations of 1 pg ml−1 and 0.1 pg ml−1 can be detected in the mixture of the proteins without any amplification technique, which represents 1000 times improvement of the sensitivity compared to the previously reported methods. The proposed method can be applied to the efficient detection of the multiple biomarkers for the diagnostic applications. Hence, the presented method is under investigation for the detection of the biomarkers in the clinical samples.
This work was supported by the Ministry of Knowledge and Economy through the International Collaborative Research Program.
Notes and references
- (a) Louisiana Department of Health and Hospitals, Office of Public Health (DHH/OPH), 2004 Louisiana Health Report Card. Louisiana Center for Health Statistics, 2005; (b) V. Kulasingam and E. P. Diamandis, Nat. Clin. Pract. Oncol., 2008, 5, 588 Search PubMed.
- J. F. Rusling, C. V. Kumara, J. S. Gutkind and V. Patele, Analyst, 2010, 135, 2496 RSC.
- (a) K. Y. Tomizaki, K. Usui and H. Mihara, ChemBioChem, 2005, 6, 782 CrossRef CAS; (b) B. Schweitzer, P. Predki and M. Snyder, Proteomics, 2003, 3, 2190 CrossRef CAS.
- (a) E. Phizicky, P. I. H. Bastiaens, H. Zhu, M. Snyder and S. Fields, Nature, 2003, 422, 208 CrossRef CAS; (b) D. Kambhampati, Protein Microarray Technology, Wiley-VCH, Verlag GmbH & Co. KGaA, 2003 Search PubMed.
- (a) J. L. Richens, E. A. M. Lunt, D. Sanger, G. McKenzie and P. O'Shea, J. Proteome Res., 2009, 8, 5103 Search PubMed; (b) H.-Y. Hsu and Y. Y. Huang, Biosens. Bioelectron., 2004, 20, 123 CrossRef CAS.
- (a) C. M. Niemeyer, T. Sano, C. L. Smith and C. R. Cantor, Nucleic Acids Res., 1994, 22, 5530 CrossRef CAS; (b) R. Wacker, H. Schröeder and C. M. Niemeyer, Anal. Biochem., 2004, 330, 281 CrossRef CAS.
- (a) R. C. Bailey, G. A. Kwong, C. G. Radu, O. N. Witte and J. R. Heath, J. Am. Chem. Soc., 2007, 129, 1959 CrossRef CAS; (b) H. Zhu and M. Snyder, Curr. Opin. Chem. Biol., 2003, 7, 55 CrossRef CAS.
- (a) C. Boozer, J. Ladd, S. Chen, Q. Yu, J. Homola and S. Jiang, Anal. Chem., 2004, 76, 6967 CrossRef CAS; (b) R. Wacker and C. M. Niemeyer, ChemBioChem, 2004, 5, 453 CrossRef CAS.
- (a) J.-M. Nam, C. S. Thaxton and C. A. Mirkin, Science, 2003, 301, 1884 CrossRef CAS; (b) M. Gullberg, S. Fredriksson, M. Taussig, J. Jarvius, S. Gustafsdottir and U. Landegren, Curr. Opin. Biotechnol., 2003, 14, 82 CrossRef CAS.
- Y. Lee, E. K. Lee, Y. W. Cho, T. Matsui, I.-C. Kang, T. Kim and H. M. Han, Proteomics, 2003, 3, 2289 CrossRef CAS.
- K. Song, S. B. Nimse, J. Kim, J. Kim, V. Nguyen, W. Eoum, V. Ta and T. Kim, Chem. Commun., 2011 10.1039/c1cc12489g.
- C. Boozer, J. Ladd, S. Chen and S. Jiang, Anal. Chem., 2006, 78, 1515 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c1cc12721g |
| This journal is © The Royal Society of Chemistry 2011 |
