 Open Access Article
Open Access Article9G DNAChip: microarray based on the multiple interactions of 9 consecutive guanines†
Keum-Soo
Song
b,
Satish
Balasaheb Nimse
a,
Joonbae
Kim
a,
Junghoon
Kim
b,
Van-Thuan
Nguyen
a,
Van-Thao
Ta
a and
Taisun
Kim
*a
aInstitute for Applied Chemistry and Department of Chemistry, Hallym University, Chuncheon, 200-702, Korea. E-mail: tskim@hallym.ac.kr; Fax: +82-33-256-3421
bBiometrix Technology, Inc. 202 BioVenture Plaza, Chuncheon, 200-161, Korea
First published on 27th May 2011
Abstract
We introduce the phenomenon of molecular recognition to immobilize oligonucleotides on AMCA slides for the production of 9G DNAChips. Facile and efficient method for the immobilization of the oligonucleotides appended with consecutive nine guanine bases is described. The 9G DNAChips shows more than 90% hybridization efficiency at 25 °C in 30 min.
The DNA chip technology has emerged as a powerful and promising tool in biological sciences.1,2 The success of DNA chips not only depends on the chemistry used for the immobilization of oligonucleotides but also depends on the good accessibility and functionality of the surface-bound oligonucleotides, density of attachment, and reproducibility of attachment chemistry.3 In recent years, the deposition method is the most frequently used method for the immobilization of oligonucleotides, where pre-synthesized oligonucleotides are covalently4 or non-covalently5 immobilized on solid surfaces (organic or inorganic). The drawbacks of this method were the decrease in hybridization efficiency with an increase in the density of the immobilized probes and the reproducibility of the immobilization method.6
The performance of DNA chips was under a shadow due to the several issues, including the probe design, the reaction conditions during spotting, the hybridization and washing conditions, the suppression of nonspecific binding, the vertical spacing between the oligonucleotides and the surface, and the lateral spacing between the immobilized oligonucleotides.7 Many research groups have noticed the unique aspect of the lateral spacing between oligonucleotides.8 The lateral spacing is not only important to make DNA chips but also to make arrays of proteins,9aptamers,10 and small molecules.11 Until now, mixed self-assembled monolayers have been generally applied to control the lateral spacing between oligonucleotides on Au electrodes.12 Park and his coworkers employed a self-assembled monolayer of nanocones to immobilize the oligonucleotides covalently and to separate them from each other.13 Whereas, another group used a self-assembled gold nanoparticle monolayer to control the spacing between the immobilized oligonucleotides.14
The approach of the multiple ionic interactions has been used to fabricate the arrays of the proteins on the calix[4]crown monolayer.15 The weak interaction was the key to allow the molecules of the proteins to be immobilized compactly without leaving any unoccupied area for further physisorption. However, in DNA chip technology, molecular recognition has never been used to immobilize oligonucleotides on the solid surface.
Recently, we have reported that the aminocalix[4]arene derivatives can efficiently recognize structurally flat and hydrophobic molecules in water.16 Based on these findings, we fabricated the monolayer of the aminocalix[4]arene derivative on the slides to obtain AMCA slides. Surprisingly, the oligonucleotides with 9 consecutive guanines (9G) can be immobilized on the AMCA slide to generate the 9G DNAChip. In this letter, we report a novel and very efficient approach to make the 9G DNAChip based on the multiple molecular interactions and its characterization. The obtained 9G DNAChips show more than 90% hybridization efficiency at 25 °C in 30 min, with very high specificity.
The preparation of the 9G DNAChip and the hybridization thereafter is briefly explained (Scheme 1). The AMCA slides are obtained by reacting the amine slide with the AMCA-1,3-dialdehyde to generate a monolayer of AMCA on the surface (see the ESI†). By spotting the solution of the oligonucleotides appended with 9 consecutive guanines (see the ESI† Table S1), the oligonucleotides can be immobilized on the AMCA slide to generate the 9G DNAChip. Cy5-labeled complementary oligonucleotides are hybridized and washed at 25 °C to evaluate the efficiency of the 9G DNAChip. The optimal condition for the effective immobilization of the oligonucleotides was determined by spotting the 20 μM of oligonucleotide probes on the AMCA slide (see the ESI† Fig. S3b). The probes used were, Probe6 (0G), Probe7 (3G), Probe8 (6G), Probe1 (9G), and Probe9 (12G). The spots were arranged to make 2 by 5 pixels and the microarray was subjected to hybridization using the Cy5-labeled probe (Cy5-T1) in hybridization buffer (pH = 7.4) and subsequent washing and drying steps, all at 25 °C.
 | ||
| Scheme 1 (A) The preparation of the 9G DNAChip and hybridization with the Cy5-labeled target DNA (Cy5-T1). | ||
The dried 9G DNAChip was scanned under a laser scanner and analyzed. The results showed that maximum immobilization was achieved for the probe appended with 9G subunits (Probe1) (Fig. 1a and b). As the number of the guanine (G) subunits in the probes was increased from 0 to 12, the fluorescence intensity increased and reached the maximum for the probe appended with 9G and decreased for the probe with 12G. This behavior of the increasing fluorescent intensity (arbitrary unit) from 0 to 9 guanine bases is correlated with the efficient immobilization of the probes with 9 guanine bases. The increase in number of guanine bases in probes increases the binding ability of the probes on the AMCA slide. Even though the probe with 12G may have higher binding capacity, it shows lower fluorescent intensity than the probe with 9G. This is explained on the basis that the number of the 12G probes immobilized on the specific area would be less than that of the 9G probes because of the three additional guanines.
 | ||
| Fig. 1 (a) A graph representing the effect of the number of the guanine bases in oligonucleotide probes on the immobilization of probes on the AMCA slides. (b) Fluorescence map obtained by the immobilization of the Probe6 (0G), Probe7 (3G), Probe8 (6G), Probe1 (9G), and Probe9 (12G) after the hybridization with Cy5-T1, PMT gain = 48. | ||
To get insight into the molecular recognition properties of AMCA monolayer on the AMCA slide, further investigation was done by immobilization of the probes appended with 9 adenine (9A, Probe3), 9 thymine (9T, Probe4), 9 guanine (9G, Probe1), and 9 cytosine (9C, Probe5) subunits, respectively, on the AMCA slide.
Four probes Probe3, Probe4, Probe1, and Probe5 were immobilized on the AMCA slides and hybridized at 25 °C using 100 fmol μl−1 of Cy5-T1. The probe with 9G (Probe1) provides the highest fluorescence intensity among all four probes (Fig. 2). The probes with 9T (Probe5) and 9C (Probe4) showed the lowest fluorescence intensity. The probe with 9A (Probe3) also showed significant fluorescence intensity but not as good as the probe with 9G (Probe1). Based on these findings, oligonucleotides with nine consecutive guanine bases were used to generate 9G DNAChips.
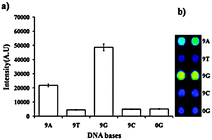 | ||
| Fig. 2 (a) A graph representing the effect of 9 consecutive DNA bases in the oligonucleotide probe on the immobilization of probes on the AMCA slides. (b) Fluorescence map obtained by immobilization of Probe3 (9A), Probe5 (9T), Probe1 (9G), Probe4 (9C), and Probe6 (0G) after the hybridization with Cy5-T1, PMT gain = 48. | ||
Recently, we reported the molecular recognition of aminocalix[4]arene derivatives with several guest molecules. The 4-picoline showed strong binding (Ka = 3.9 × 104) with the host, while benzoic acid did not show any affinity. To provide evidence for the immobilization of the probes based on molecular recognition, the immobilization of Probe1 in the presence of benzoic acid and 4-picoline was carried out and analyzed after hybridization.
The 20 μM of the Probe1 was immobilized on the AMCA slide in the presence of the 4-picoline or benzoic acid with increasing concentrations from 0–100 mM. After the hybridization with Cy5-T1, the 9G DNAChip was scanned and analyzed. The molecules of the 4-picoline moderately prevented the immobilization of the Probe1 on the AMCA slides, while benzoic acid did not show any effect on the immobilization (Fig. 3). From these results, it is clear that the 4-picoline competes with the 9 guanines for the cavities of the AMCA during the immobilization process. Based on these observations, the immobilization efficiency and kinetics of the probes appended with 9G were studied in detail (see the supporting information†). In 4 h, the immobilization density of 6.3 pmol cm−2 was achieved using 20 μM of Cy5-Probe 1 (see the ESI† Fig. S4, Table S2).
 | ||
| Fig. 3 (a) Competition of guest molecules with 9G appended probes (Probe1) during immobilization on the AMCA slide. (b) Fluorescence map obtained by immobilization of Probe1 in presence of various concentrations of guests after hybridization with Cy5-T1, PMT gain = 48. | ||
The drawback of most of the reported immobilization methods was the decrease in hybridization efficiency with the increase in concentration of immobilized probes. This was explained on the basis that overloading the surface with probes might cause a crowding effect that could lower the accessibility of the surface-bound probes.17
The 9G DNAChip shows the hybridization efficiencies (Fig. 4) of the 44%, 65%, and 93% for the probe with the immobilization concentrations of 2.5 μM, 5.0 μM, and 10 μM, respectively (for details on calculation of the hybridization efficiency please see the ESI†Fig. 3). Interestingly, the hybridization efficiency increases with the increase in the concentration of the immobilized probe. At a concentration of 10 pmol μl−1, the hybridization efficiency reaches 93% and does not increase significantly, even the concentration increases up to 33 pmol/μl. This can be explained on the basis of the excellent accessibility to the immobilized probes due to the lateral spacing provided by 9 guanines.
 | ||
| Fig. 4 Hybridization efficiency obtained by comparing (c) to (b). (a) A graph representing the hybridization efficiency obtained by comparing (c) to (b). (b) The fluorescence image of the immobilized Cy5-Probe1 at the concentration of 2.5, 5, 10, 20, and 33 μM. (c) The fluorescence image obtained by the immobilization of Probe1 at the concentration of 2.5, 5, 10, 20, and 33 μM and the hybridization with Cy5-T1 (0.1 μM) for 30 min, and the figures to the left indicate the percentage hybridization efficiency as compared to b), PMT gain = 48. | ||
To investigate the sensitivity of the 9G DNAChip, four different 9G probes were spotted on the AMCA slide to obtain the 9G DNAChip. Probe1, Probe1-m1, Probe1-m2, and Probe2 have zero, one, two mismatches, and noncomplementary to Cy5-T1, respectively. The 9G DNAChip was hybridized with Cy5-T1 at 25 °C for 30 min, washed and analyzed (see the ESI† Fig. S6a). The fluorescence images show that there is a dramatic difference in the intensity between the matched and the mismatched sequences. The perfectly matched sequence (Probe1) gave the maximum intensity. As compared to the spots with Probe1, the spots for probes having one (Probe1-m1) and two (Probe1-m2) mismatches showed 5.5 and 20 times decreased fluorescence intensity, respectively. There were no measurable hybridization signals from the spots of the noncomplementary (Probe2) probe, indicating that the nonspecific hybridization did not occur. The excellent specificity is considered due to the lateral spacing endowed by the 9 guanines. The exact geometry of the immobilized 9G probes on the AMCA slide is under investigation and will be reported elsewhere.
Moreover, the efficiency of the 9G DNAChip is attributed to the AMCA slide. The monolayer of the AMCA molecules on the AMCA slide produces a slightly hydrophobic surface (see the ESI† Fig. S2b, contact angle θ = 68.6°) as compare to bare slide glass, which enables the uniformity of the spots by prohibiting the immobilization solution to spread. The fluorescence map (see the ESI† Fig. S6b) depicts the quality of the AMCA slide in terms of the spot morphology, the spot homogeneity, and the signal reproducibility.
In conclusion, based on our knowledge on DNA chip technology, we are the first to introduce the phenomenon of molecular recognition to immobilize the oligonucleotides for the production of DNA chips. The 9G probes are immobilized by the multiple interactions of the 9 consecutive guanines on the AMCA monolayer. The lateral spacing between the immobilized probes provides the high accessibility leading to the more than 90% hybridization efficiency in 30 min at 25 °C. Moreover, the 9 consecutive guanines can be easily added to the oligonucleotide probes during their synthesis. The excellent properties shown by the 9G DNAChip enables it to be a powerful and promising tool for the biotechnology. We are currently working on the applications of the 9G DNAChip for the detection of the biomolecules and biomolecular complexes.
This research was financially supported by the Ministry of Education, Science and Technology (MEST) and Korea Industrial Technology Foundation (KOTEF) through the Human Resource Training Project for Regional Innovation.
Notes and references
- (a) G. Ramsay, Nat. Biotechnol., 1998, 16, 40 CrossRef CAS; (b) J. R. Pollock, C. M. Perou, A. A. Alizadeh, M. B. Eisen, A. Pergamenschikov, C. F. Williams, S. S. Jeffery, D. Botstein and P. O. Brown, Nature Genet., 1999, 23, 41.
- (a) A. N. Drobyshev, N. Mologina, V. Shick, D. Pobedimskaya, G. Yershov and A. Mirzabekov, Gene, 1997, 188, 45 CrossRef CAS; (b) R. E. Sosnowskii, E. Tu, W. Butler, J. O'Connell and M. Heller, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 1119 CrossRef CAS.
- (a) A. Sassolas, B. D. Leca-Bouvier and L. J. Blum, Chem. Rev., 2008, 108, 109 CrossRef CAS; (b) X. Zhao, S. Nampalli, A. J. Serino and S. Kumar, Nucleic Acids Res., 2001, 29, 955 CrossRef CAS.
- (a) P. Kumar, S. K. Agrawal, A. Misra and K. C. Gupta, Bioorg. Med. Chem. Lett., 2004, 14, 1097 CrossRef CAS; (b) B. Joos, H. Kuster and R. Cone, Anal. Biochem., 1997, 247, 96 CrossRef CAS.
- G. Nonglaton, I. O. Benitez, I. Guisle, M. Pipelier, J. Leger, D. Dubreuil, C. Tellier, D. R. Talham and B. Bujoli, J. Am. Chem. Soc., 2004, 126, 1497 CrossRef CAS.
- (a) N. E. Broude, K. Woodward, R. Cavallo, C. R. Cantor and D. Englert, Nucleic Acids Res., 2001, 29, e92 CAS; (b) P. Kumar and K. C. Gupta, Bioconjugate Chem., 2003, 14, 507 CrossRef CAS.
- M. Dufva, Biomol. Eng., 2005, 22, 173 CrossRef CAS.
- (a) B. J. Hong, S. J. Oh, T. O. Youn, S. H. Kwon and J. W. Park, Langmuir, 2005, 21, 4257 CrossRef CAS; (b) K. M. A. Rahman, C. J. Durning, N. J. Turro and D. A. Tomalia, Langmuir, 2000, 16, 10154 CrossRef CAS.
- G. MacBeath and S. L. Schreiber, Science, 2000, 289, 1760 CAS.
- H. Xu, X. Mao, Q. Zeng, S. Wang, A. Kawde and G. Liu, Anal. Chem., 2009, 81, 669 CrossRef CAS.
- T. Kodadek, Chem. Biol., 2001, 8, 105 CrossRef CAS.
- A. W. Peterson, L. K. Wolf and R. M. Georgiadis, J. Am. Chem. Soc., 2002, 124, 14601 CrossRef CAS.
- Y. Choi, C. W. Yoon, H. D. Lee, M. Park and J. W. Park, Chem. Commun., 2004, 1316 RSC.
- T. Ohashi, K. Mawatari and T. Kitamori, Biomicrofluidics, 2010, 4, 032209 CrossRef.
- Y. Lee, E. K. Lee, Y. W. Cho, T. Matsui, I. Kang, T. Kim and M. H. Han, Proteomics, 2003, 3, 2289 CrossRef CAS.
- (a) S. B. Nimse, K. Song, C. Jung, W. Eoum and T. Kim, Bull. Korean Chem. Soc., 2009, 30, 1247 CrossRef CAS; (b) S. B. Nimse, V. Nguyen, J. Kim, H. Kim, K. Song, W. Eoum, C. Jung, V. Ta, S. R. Seelam and T. Kim, Tetrahedron Lett., 2010, 51, 2840 CrossRef CAS; (c) S. B. Nimse, K. Song, J. Kim, H. Kim, V. Nguyen, W. Eoum, C. Jung, V. Ta and T. Kim, Tetrahedron Lett., 2010, 51, 6156 CrossRef CAS.
- S. Mahajan, P. Kumar and K. C. Gupta, Bioconjugate Chem., 2006, 17, 1184 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Synthesis and characterization of the AMCA-1,3-dialdehyde, experimental procedure for preparation of the AMCA slide, the 9G DNAChip and hybridization. See DOI: 10.1039/c1cc12489g |
| This journal is © The Royal Society of Chemistry 2011 |
