 Open Access Article
Open Access ArticleChemoenzymatic synthesis of sialooligosaccharides on arrays for studies of cell surface adhesion†‡
Róbert
Šardzík
a,
Ritu
Sharma
b,
Sara
Kaloo
a,
Josef
Voglmeir
a,
Paul R.
Crocker
*b and
Sabine L.
Flitsch
*a
aManchester Interdisciplinary Biocentre & School of Chemistry, The University of Manchester, 131 Princess Street, Manchester, M1 7DN, UK. E-mail: sabine.flitsch@manchester.ac.uk; Fax: +44 (0)161 2751311; Tel: +44 (0)161 3065172
bCollege of Life Sciences, University of Dundee, Dow Street, Dundee, DD1 5EH, UK
First published on 5th April 2011
Abstract
Sialooligosaccharides were generated by direct enzymatic glycosylation on arrays and the resulting surfaces were suitable for the study of carbohydrate-specific cell adhesion.
Sialic acid conjugates play a dominant role in many cellular recognition processes in health and disease. They are frequently exploited by viruses and bacteria as sites of attachment to cells and extracellular proteins and they can function as recognition determinants for host cell lectins, including the selectin and siglec families involved in inflammatory and immune responses.1 Pioneering work over the last ten years has established glycan arrays as central tools to understanding many diverse carbohydrate–protein interactions in cells and organisms.2,3 However, both the complex preparation of oligosaccharide arrays and the lengthy biochemical generation of the labelled proteins have precluded more genome wide investigations of sialic acid binding proteins beyond first examples. Here we present some dramatic shortcuts to the present methods of analysis with three key components to our overall strategy: Firstly, the complex sialooligosaccharides are generated directly on the array using chemoenzymatic synthesis. Secondly, the efficiency of synthesis is monitored in situ using MALDI-ToF mass spectrometry. Thirdly, the sialooligosaccharides are directly interrogated by recombinant mammalian cells expressing siglecs on their surfaces.
The overall strategy is shown in Fig. 1. Self-assembled monolayers (SAMs) of alkanethiols on gold surfaces provide well established platforms for the study of enzymatic reactions and protein–ligand and cell–ligand interactions4–7 and are ideally suited for our studies.8 We have previously shown how glycan arrays on SAM-gold can be used to study the specificity of glycosyltransferases by using MALDI-ToF MS9 as a label free read-out method. Importantly, we have been able to show that the reactions can be quantitative, thus allowing for the direct enzymatic synthesis and analysis of saccharides on array surfaces, without the need for multistep solution phase synthesis and subsequent linkage to array.6,7
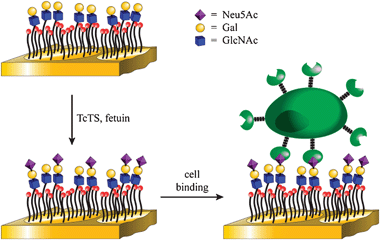 | ||
| Fig. 1 Overall strategy for the chemoenzymatic synthesis and cell-based detection of sialooligosaccharides. | ||
A concern for the synthesis of sialooligosaccharides was the use of harsh MALDI techniques employed in the analysis of enzymatic reactions on the arrays, given the known sensitivity of sialosides to this technique.10 Indeed, when we first attempted the enzymatic transfer of sialic acid from fetuin onto immobilised lactose411 (Fig. 2) using trans-sialidase from Trypanosoma cruzi (TcTS)12 analysis by previously published6,7MALDI-ToF MS analysis failed. Only peaks corresponding to the starting material 4 were seen in the MS spectra which could have been due to lack of enzymatic activity or cleavage of labile sialic acid groups.10,13
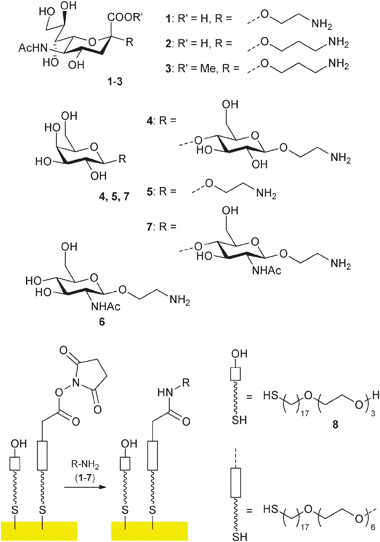 | ||
| Fig. 2 Immobilisation of glycosylamines1–7 on SAMs/gold surfaces. | ||
The degradation of sialosides during mass spectrometric analysis was further investigated with immobilised 2-aminoethyl and 3-aminopropyl glycoside of Neu5Ac (1 and 2 respectively) together with methyl ester 3.11
We first focussed on optimising the MALDI matrix and found 2,4,6-trihydroxyacetophenone (THAP) to be best in our hands, although significant degradation was still observed for 1 (See supplementary information, Fig. S1). Interestingly, with the propyl compound 2 only a very minor and with the ester3 no fragmentation was observed during the MS experiments (Fig. S2, S3 in supporting information), indicating that the linker plays a significant role in stability and that methylation increases stability of the sialoside.
The stability of the methyl ester prompted us to investigate methylation protocols, which are well developed in solution phase13 but had not been used directly on the SAM-gold surfaces. In our hands the most successful procedure proved to be the methyl ester formation using MeI in DMSO as described by Powell and Harvey.13 The on-chip conversion of the acid 2 to its sodium salt followed by treatment with MeI in DMSO afforded very similar spectra to those of the ester3 (Fig. S4, S5 in supporting information). Alternatively, permethylation of all sialo structures 1, 2 and 3 with MeI and sodium dimsyl in DMSO as described by Hakomori14 gave strong signals, referring to permethylated aminoethyl- and aminopropyl Neu5Ac respectively. (Fig. S6–S8 in supporting information).
With a reliable protocol for on-chip analysis of sialooligosaccharides in hand, enzymatic sialylation (Fig. 3) with trans-sialidase from Trypanosoma cruzi (TcTS) and fetuin as donor12 was performed on immobilised lactose derivative 4, galactose5 and also N-acetyl-lactosamine 7, the latter generated enzymatically from N-acetyl glucosamine 6 using bovine β1,4-galactosyltransferase (β1,4-GalT) as described before.6 It was now indeed possible to detect all sialylated structures (Fig. 3, S9–S12 in supporting information). The mass spectra suggested that sialylation had proceeded in reasonable yield (estimated about 50% from peak heights in the MS). This is comparable to solution phase yields observed for the TcTS.12
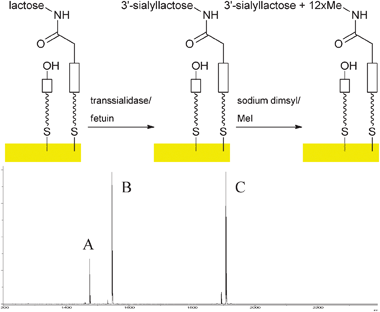 | ||
| Fig. 3 Enzymatic sialylation of immobilised lactoside on gold surfaces and MALDI ToF MS analysis after permethylation (A: m/z 1474, permethylated sialyl lactoside; B: m/z 1545, heterodimer of starting material lactoside with 8; C: m/z 1906, heterodimer of permethylated sialyl lactoside with 8). | ||
With the sialolactoside in hand we investigated the binding of siglecs to our surfaces. Many carbohydrate-binding proteins are membrane associated cell surface receptors. Analysis of their carbohydrate binding specificities with glycan arrays usually involves their conversion to soluble proteins which are expressed in heterologous cells and then purified, labelled and crosslinked to obtain high avidity binding.3 An alternative, more physiological, approach would be to measure array binding using fluorescentlylabelled cells expressing these proteins on the plasma membrane (third step in Fig. 1). Such experiments have been attempted with glass microarrays, but have produced limited results.15,16 Self-assembled monolayers on gold, however, seem particularly suited to such cell binding assays because of low levels of non-specific cellular adhesion reported for arrays containing cell-adhesion peptides.8,17 In addition, the SAM-gold surfaces appear to be able to present ligands at high densities which might be important for lectins that are involved in polyvalent binding.18 Thus we investigated the binding of CHO cells expressing sialoadhesin, a member of the siglec family, on the cell surface.1,19 The sialyllactoside ligands were presented at different densities in array format, which was achieved by surface dilution of functionalised SAMs with triethylene glycol groups using 8.4,6,7,20 The CHO cells were labelled with cell-permeable dye (CFSE; carboxyfluorescein diacetate succinimidyl ester) and were applied to our sialylated array surfaces. Fig. 4 shows that fluorescence intensities of surfaces containing sialyllactose are significantly stronger than those of surfaces containing lactose and that cell binding is dependent on the density of sugar on the surface (lowest at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 sugar
9 sugar![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethylene glycol8). The bottom panel shows the negative control containing lactoside 4 before sialylation and suggests that very little non-specific adhesion is observed under the assay conditions.
ethylene glycol8). The bottom panel shows the negative control containing lactoside 4 before sialylation and suggests that very little non-specific adhesion is observed under the assay conditions.
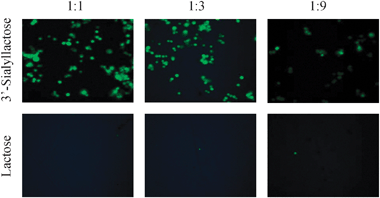 | ||
| Fig. 4 Binding of CFSE-labelled CHO cells expressing sialoadhesin to arrays displaying 3′-sialyllactose (top panel) and lactose (bottom panel) on gold at three surface dilutions. | ||
In conclusion, we have shown that sialooligosaccharides can be generated directly on array surfaces using enzymatic synthesis in good yields, thus providing much faster access to these glycan ligands than solution synthesis combined with immobilisation. Because of the on-chip synthesis, the analysis by mass spectrometry provides an additional and essential step of quality control to confirm composition of the array surface. We have also shown that the SAM-gold platform is an ideal surface for studying cellular adhesion through protein–carbohydrate interactions with low background observed and should provide a useful tool for the study of recombinant cell-surface receptors of glycans and other ligands.
This work was supported by the BBSRC, RCUK, The European Commission, The Wellcome Trust Senior Fellowship (WT081882) and The Royal Society.
Notes and references
- P. R. Crocker, J. C. Paulson and A. Varki, Nat. Rev. Immunol., 2007, 7, 255 CrossRef CAS; S. J. McMillan and P. R. Crocker, Carbohydr. Res., 2008, 343, 2050 CrossRef CAS; M. Sperandio, C. A. Gleissner and K. Ley, Immunol. Rev., 2009, 230, 97 CrossRef CAS.
- T. Horlacher and P. H. Seeberger, Chem. Soc. Rev., 2008, 37, 1414 RSC; N. Laurent, J. Voglmeir and S. L. Flitsch, Chem. Commun., 2008, 4400 RSC; J. Stevens, O. Blixt, J. C. Paulson and I. A. Wilson, Nat. Rev. Microbiol., 2006, 4, 857 Search PubMed; J. Voglmeir, R. Sardzik, M. J. Weissenborn and S. L. Flitsch, OMICS, 2010, 14, 437 CrossRef CAS.
- Y. Liu, T. Feizi, M. A. Carnpanero-Rhodes, R. A. Childs, Y. N. Zhang, B. Muiioy, P. G. Evans, H. M. I. Osborn, D. Otto, P. R. Crocker and W. C. Chai, Chem. Biol., 2007, 14, 847 CrossRef CAS.
- B. T. Houseman, J. H. Huh, S. J. Kron and M. Mrksich, Nat. Biotechnol., 2002, 20, 270 CrossRef CAS; B. T. Houseman and M. Mrksich, Angew. Chem., Int. Ed., 1999, 38, 782 CrossRef CAS; M. Mrksich, G. B. Sigal and G. M. Whitesides, Langmuir, 1995, 11, 4383 CrossRef.
- R. Karamanska, J. Clarke, O. Blixt, J. I. MacRae, J. Q. Q. Zhang, P. R. Crocker, N. Laurent, A. Wright, S. L. Flitsch, D. A. Russell and R. A. Field, Glycoconjugate J., 2008, 25, 69 CrossRef CAS.
- Z. L. Zhi, N. Laurent, A. K. Powell, R. Karamanska, M. Fais, J. Voglmeir, A. Wright, J. M. Blackburn, P. R. Crocker, D. A. Russell, S. Flitsch, R. A. Field and J. E. Turnbull, ChemBioChem, 2008, 9, 1568 CrossRef CAS.
- N. Laurent, R. Haddoub, J. Voglmeir, S. C. C. Wong, S. J. Gaskell and S. L. Flitsch, ChemBioChem, 2008, 9, 2592 CrossRef CAS; N. Laurent, J. Voglmeir, A. Wright, J. Blackburn, N. T. Pham, S. C. C. Wong, S. J. Gaskell and S. L. Flitsch, ChemBioChem, 2008, 9, 883 CrossRef CAS.
- R. T. Petty, H. W. Li, J. H. Maduram, R. Ismagilov and M. Mrksich, J. Am. Chem. Soc., 2007, 129, 8966 CrossRef CAS.
- M. Mrksich, ACS Nano, 2008, 2, 7 CrossRef CAS; N. Nagahori and S. I. Nishimura, Chem.–Eur. J., 2006, 12, 6478 CrossRef CAS.
- D. J. Harvey, Mass Spectrom. Rev., 2008, 27, 125 CrossRef CAS.
- R. Sardzik, G. T. Noble, M. J. Weissenborn, A. Martin, S. J. Webb and S. L. Flitsch, Beilstein J. Org. Chem., 2010, 6, 699 Search PubMed.
- M. F. Amaya, A. G. Watts, I. Damager, A. Wehenkel, T. Nguyen, A. Buschiazzo, G. Paris, A. C. Frasch, S. G. Withers and P. M. Alzari, Structure, 2004, 12, 775 CrossRef CAS; V. L. Campo, I. Carvalho, C. Da Silva, S. Schenkman, L. Hill, S. A. Nepogodiev and R. A. Field, Chem. Sci., 2010, 1, 507 RSC.
- A. K. Powell and D. J. Harvey, Rapid Commun. Mass Spectrom., 1996, 10, 1027 CrossRef CAS.
- S. I. Hakomori, J. Biochem., 1964, 55, 205 CAS.
- L. Nimrichter, A. Gargir, M. Gortler, R. T. Altstock, A. Shtevi, O. Weisshaus, E. Fire, N. Dotan and R. L. Schnaar, Glycobiology, 2004, 14, 197 CrossRef CAS.
- O. Blixt, S. F. Han, L. Liao, Y. Zeng, J. Hoffmann, S. Futakawa and J. C. Paulson, J. Am. Chem. Soc., 2008, 130, 6680 CrossRef CAS.
- K. Mougin, M. B. Lawrence, E. J. Fernandez and A. C. Hillier, Langmuir, 2004, 20, 4302 CrossRef CAS; D. Peelen, V. Kodoyianni, J. Lee, T. Zheng, M. R. Shortreed and L. M. Smith, J. Proteome Res., 2006, 5, 1580 CrossRef CAS.
- T. K. Lindhorst, K. Bruegge, A. Fuchs and O. Sperling, Beilstein J. Org. Chem., 2010, 6, 801 Search PubMed; N. P. Pera, H. M. Branderhorst, R. Kooij, C. Maierhofer, M. van der Kaaden, R. M. J. Liskamp, V. Wittmann, R. Ruijtenbeek and R. J. Pieters, ChemBioChem, 2010, 11, 1896 CrossRef.
- P. R. Crocker, S. Kelm, A. Hartnell, S. Freeman, D. Nath, M. Vinson and S. Mucklow, Biochem. Soc. Trans., 1996, 24, 150 CAS.
- R. G. Chapman, E. Ostuni, L. Yan and G. M. Whitesides, Langmuir, 2000, 16, 6927 CrossRef CAS; J. C. Love, L. A. Estroff, J. K. Kriebel, R. G. Nuzzo and G. M. Whitesides, Chem. Rev., 2005, 105, 1103 CrossRef CAS.
Footnotes |
| † This article is part of the ‘Glycochemistry and glycobiology’ web-theme issue for ChemComm. |
| ‡ Electronic supplementary information (ESI) available. See DOI: 10.1039/c1cc10745c |
| This journal is © The Royal Society of Chemistry 2011 |
