Cellular binding of nanoparticles in the presence of serum proteins†‡
Gerard W.
Doorley
and
Christine K.
Payne
*
School of Chemistry and Biochemistry and Petit Institute for Bioengineering and Bioscience, Georgia Institute of Technology, 901 Atlantic Drive, Atlanta, Georgia 30332, USA. E-mail: christine.payne@chemistry.gatech.edu; Fax: +1 404-385-6057; Tel: +1 404-385-3125
First published on 1st October 2010
Abstract
Cellular binding of cationic nanoparticles in the presence of serum proteins was probed with two-colour fluorescence microscopy. Cationic nanoparticles associate with serum proteins in solution and bind to the cell surface as a single anionic complex. Displacement of serum proteins from the nanoparticles was found to be protein dependent.
Nanoparticles (NPs) have important biomedical applications ranging from the treatment of human disease with gene therapy to understanding basic cellular functions with fluorescent probes. NPs for use in cellular applications have been synthesized from materials that include polymers,1–3 semiconductors,4carbon,5 and noble metals.6,7 Common to all of these materials is the need to functionalize the NP for cellular binding, internalization, and targeting.
Cationic peptides, polymers, and lipids are the ligands most commonly used to initiate the cellular binding and endocytic uptake of NPs.1,8–14Endocytosis can be highly specific with a receptor on the cell surface recognizing a specific ligand and initiating the internalization of the ligand into an endocytic vesicle.15 In comparison, the cationic ligands conjugated to NPs are often synthetic and lack dedicated receptors on the cell surface. Although the endocytic uptake of cationic ligand–NPs is typically described as non-specific, there are key elements in this pathway that appear to be common to many of the cationic NPs. Many of the cationic ligands used for the functionalization of NPs are dependent on proteoglycans for binding to the cell surface.16–24Proteoglycans consist of a central protein bound to multiple sugar side chains.25 The highly-sulfated sugar side chains of the proteoglycans provide dense regions of negative charge. Previous studies have shown that removal of proteoglycans or their sulfate groups will inhibit endocytosis of cationic NPs.21,24
While binding of cationic NPs to anionic proteoglycans on the cell surface is conceptually straightforward, recent work characterizing the surface of NPs in the presence of serum proteins presents a more complex picture of NP–cell interactions.26,27 Medium in which cells are cultured, referred to as cell culture medium, typically consists of two components. The medium alone is an aqueous solution of amino acids, vitamins, inorganic salts, and glucose. For cell culture this medium is supplemented with serum, a protein solution separated from the whole blood of cows, pigs, horses, or other animals. While the medium is a carefully controlled solution, the serum is a highly complex and varied mixture of proteins.28
Results with a range of different NPs including gold nanorods,29Al2O3,30silica,31TiO2, carbon black, fullerol,32polymers,31–34 and a correspondingly broad range of techniques including zeta potential measurements,29,30,32dynamic light scattering,29,31,32 differential centrifugal sedimentation,31transmission electron microscopy,31isothermal titration calorimetry,33surface plasmon resonance,33 and size-exclusion chromatography33 suggest a model in which NPs, both cationic and anionic, rapidly bind a mixture of proteins present in serum resulting in an anionic NP that essentially presents a surface of serum proteins to the cell surface.
Of particular interest is how this serum protein–NP complex interacts with the cell surface. Specifically, does the serum protein bind to the cell with the NP or is it displaced at the cell surface by a higher affinity membrane protein. Previous work with N-isopropylacrylamide NPs in serum has shown that human serum albumin is the initial binder, but it is replaced by higher affinity proteins in solution.34 It is possible that a similar effect occurs on the cell surface with a membrane protein displacing the serum protein.
We sought to understand how NPs interact with the cell surface in the presence of serum proteins. We focused on an essential step in this process, binding of NPs to cells following exposure to the serum proteins found in cell culture medium. These experiments were carried out using simple NPs; cationic, amine-modified, 200 nm, polystyrene spheres (FluoSpheres, Invitrogen) with no additional ligands. We first measured the effective surface charge of the NPs in the presence and absence of serum proteins (Fig. 1). The NPs have a positive zeta potential of +40 mV in water. Subsequent measurements were carried out in Minimum Essential Medium (MEM) and MEM supplemented with 10% fetal bovine serum (FBS). MEM supplemented with 10% FBS is a common cell culture medium for multiple cell lines including the monkey kidney cells (BS-C-1, ATCC) used in these experiments. Zeta potential measurements in the presence of MEM show a decreased, but still positive, zeta potential (Fig. 1). A solution of 1% MEM in water shows a slight decrease, from +40 mV to +27 mV. Higher concentrations of MEM show that the zeta potential plateaus at +5 mV. The addition of FBS in MEM results in a net negative charge on the NP. A 0.01% solution of FBS results in an effective surface charge of −14 mV on the NP. Similar zeta potentials were measured for FBS in water (Fig. S1, ESI‡).
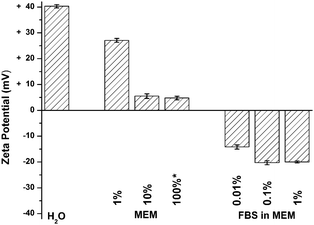 | ||
| Fig. 1 Zeta potential measurements of NPs in water, MEM (v/v%, see ESI‡) and FBS (v/v%) in MEM. FBS measurements were made in MEM supplemented with 10% FBS, diluted in water. Volume percents reflect the amount of FBS in solution. The amount of MEM is 10× the amount of FBS such that 1% FBS in MEM corresponds to a 10% MEM solution. | ||
The above results demonstrate that serum proteins bind to cationic NPs in solution resulting in anionic FBS–NPs. We next sought to determine if serum proteins remain bound to the NP on the cell surface, possibly due to a more complex interaction with proteoglycans,35 or if the serum proteins dissociate upon binding to the cell surface. To test these two possibilities it is necessary to observe both the FBS and NP as they interact with the plasma membrane. Multi-colour fluorescence imaging is ideal for this application as it allows one to image multiple fluorophores simultaneously as they interact with live cells. The amine-modified NPs described above are fluorescent with an absorption at 505 nm and an emission at 515 nm. FBS or bovine serum albumin (BSA) was labeled with AlexaFluor647 (AF647, 650 nm absorption, 665 nm emission, Invitrogen, A20006) according to the manufacturer's instructions. In brief, 125 μM BSA was incubated with 640 μM AF647 for 1 hour at pH 8. The reaction was stopped with the addition of a 1000-fold excess of hydroxylamine, which also prevented the reaction of AF647 with the amine-modified NPs (Fig. S2, ESI‡). Excess AF647 was removed with a Nap5 size exclusion column (GE Healthcare). Final concentrations of protein and AF647 were measured with a UV-Vis spectrophotometer (DU800, Beckman Coulter, Fullerton, CA, USA). The NPs and fluorescently-labeled FBS (AF647-FBS) were incubated at room temperature for 20 minutes and then added to BS-C-1 cells cultured in 35 mm glass-bottomed Petri dishes (MatTek, Ashland, MA, USA). The cells were placed in MEM alone prior to the addition of the AF647-FBS and NPs. Cells were incubated with the AF647-FBS and NPs for 10 minutes at room temperature to allow binding to the plasma membrane. The cells were then rinsed with phenol-red free MEM and imaged with an inverted microscope (Olympus IX71, Center Valley, PA, USA) with a 60×, 1.20 N.A., water immersion objective (Olympus).
Two-colour images taken after the 10 minute incubation show 100% colocalization of the AF647-FBS and NPs on the cell surface (Fig. 2A), suggesting that FBS and NPs bind to the cell surface as an FBS–NP complex. FBS is a complex mixture of different proteins. To have a better defined ligand for quantitative measurements, BSA, the major component of plasma,28 was labeled with AF647 using the method described above. Like FBS, the addition of BSA to cationic NPs results in anionic BSA–NPs in solution (Fig. S1, ESI‡). Two-colour imaging was repeated with AF647-BSA and NPs (Fig. 2B) at a maximum ratio of 8000 BSA molecules to each NP. This number of BSA molecules is on the order of the amount calculated to form a monolayer on the NP and does not affect the size of the NP (Fig. S3, ESI‡). Identical results of 100% colocalization were obtained when cells were imaged immediately following binding to the cell surface. This demonstrates that serum proteins and cationic NPs bind as a complex on the plasma membrane.
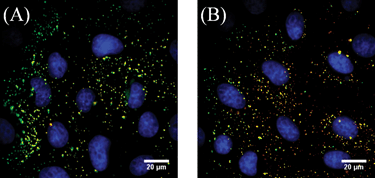 | ||
| Fig. 2 Fluorescence images of NPs (green) and serum proteins (red) bound to BS-C-1 cells after a 10 min incubation at room temperature. Colocalized signals appear yellow. Nuclei are stained with DAPI (blue). (A) FBS–NPs. (B) BSA–NPs. Unmerged images are shown in Fig. S4 (ESI‡). | ||
We then considered how the complex between the serum protein and NP changes over time to determine if the serum proteins were displaced from the NP after binding to the cell surface. This was tested by incubating FBS–NPs and BSA–NPs for 18 hours at 37 °C in the presence of MEM supplemented with 10% unlabeled FBS (Fig. 3 and Fig. S5, ESI‡). These relatively large, 200 nm NPs, without an endocytic ligand, were chosen specifically for their inability to enter the cell, allowing us to study binding as an isolated event. While the NPs lack functionalization with a traditional endocytic ligand, BSA can act as an endocytic ligand36–38 and internalization after 18 hours was measured to ensure that the NPs remained on the cell surface. Minimal (<5%) endocytic uptake was observed following the 18 hour incubation (ESI‡).
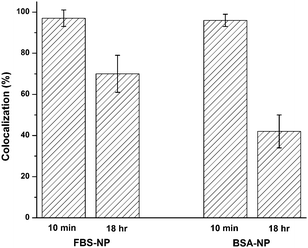 | ||
| Fig. 3 Colocalization of NP and AF647 signals for FBS–NPs and BSA–NPs at 10 minutes and 18 hours after addition to cells. The cells were fixed in 4% formaldehyde at the 18 hour time point. Error bars represent the standard deviation of 9–11 cells with an average of 69 NPs/cell. Representative images for both FBS and BSA are shown in Fig. S5 (ESI‡). | ||
Despite the excess of unlabeled FBS that could exchange with the AF647-FBS on the NP surface, only a 27% decrease in colocalization of AF647-FBS and NPs was observed after 18 hours. A greater effect was observed for AF647-BSA with a 54% decrease in colocalization after the 18 hour incubation. Lack of observed colocalization between a NP and AF647-BSA indicates that greater than 99.9% of the AF647-BSA has been displaced from the NP based on our limits of detection for AF647-BSA (Fig. S6, ESI‡). In solution, in the absence of cells, the zeta potential and hydrodynamic diameter of FBS–NPs remain stable over an 18 h incubation at 37 °C. The zeta potential undergoes a slight increase, from −14 to −8 mV, but remains negative (Fig. S7, ESI‡).
The importance of serum protein–NP interactions in biological applications is becoming increasingly well-recognized.26,27,31 The research described above confirms that cationic NPs bind to serum proteins in solution resulting in anionic protein–NPs (Fig. 1) as has been observed previously for a range of NPs including surfactant- and polymer-coated gold nanorods,29 colloidal Al2O3,30 and similar amine-modified polystyrene NPs.32 The major contribution of this research is the direct observation of the first step in the NP–cell interaction; binding of the NP to the cell surface (Fig. 2). Two-colour fluorescence imaging using FBS and BSA labeled with a red fluorophore and NPs labeled with a green fluorophore makes it possible to determine if the serum protein remains bound to the NP upon binding to the cell surface. For both FBS and BSA, 100% colocalization between the serum protein and the NP was observed on the cell surface demonstrating that FBS and BSA remain bound to the NP as it binds to the plasma membrane. Over an 18 hour incubation, relatively little FBS is displaced from the NP surface while BSA is more readily displaced (Fig. 3). This is in good agreement with previous work with serum proteins and polymer NPs that showed human serum albumin was displaced by higher affinity apolipoproteins that are also components of plasma.34 As our labeling method will label multiple protein components of FBS, it is possible that higher affinity proteins remain bound while albumin is displaced. In the case of AF647-FBS, both the higher affinity proteins and albumin are labeled and indistinguishable with fluorescence measurements. In comparison, AF647-BSA is displaced by either free, unlabeled, FBS in solution or proteins on the cell surface.
This work demonstrates that NP binding to the cell surface occurs as a serum protein–NP complex. The specific interactions that lead to cellular binding, as well as the influence of the serum proteins on the endocytic pathway of the NP, remain to be determined. In terms of binding, recent work has shown that cationic glycopolymer–DNA NPs interact with proteoglycans on the cell surface independently of charge on the NP,35 suggesting a more complex interaction than electrostatics alone. Additionally, previous work has shown that certain membrane-associated proteins have a higher affinity for albumin bound to gold NPs than albumin alone.36 These, or similar proteins, may also play a role in the cellular binding of the serum protein–NP complex. It is hoped that a better understanding of the serum protein–NP interaction, especially on the cell surface, will aid in the rational design of ligands used for NP and gene delivery in protein-rich environments, such as those found in in vivo applications.
The authors thank the NIH Director's New Innovator Award (to C.K.P., 1DP2OD006470) for financial support and Prof. Nils Kröger and Dr. Nicole Poulsen for use of the Zetasizer.
Notes and references
- O. Boussif, F. Lezoualc'h, M. A. Zanta, M. D. Mergny, D. Scherman, B. Demeneix and J. P. Behr, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 7297 CAS.
- J. J. Green, R. Langer and D. G. Anderson, Acc. Chem. Res., 2008, 41, 749 CrossRef CAS.
- C. Wu, B. Bull, C. Szymanski, K. Christensen and J. McNeill, ACS Nano, 2008, 2, 2415 CrossRef CAS.
- A. P. Alivisatos, Science, 1996, 271, 933 CrossRef CAS.
- L. Cao, X. Wang, M. J. Meziani, F. S. Lu, H. F. Wang, P. J. G. Luo, Y. Lin, B. A. Harruff, L. M. Veca, D. Murray, S. Y. Xie and Y. P. Sun, J. Am. Chem. Soc., 2007, 129, 11318 CrossRef CAS.
- J. Zheng, P. R. Nicovich and R. M. Dickson, Annu. Rev. Phys. Chem., 2007, 58, 409 CrossRef CAS.
- S. Eustis, H. Y. Hsu and M. A. El-Sayed, J. Phys. Chem. B, 2005, 109, 4811 CrossRef CAS.
- S. R. Schwarze, K. A. Hruska and S. F. Dowdy, Trends Cell Biol., 2000, 10, 290 CrossRef CAS.
- I. M. Kaplan, J. S. Wadia and S. F. Dowdy, J. Controlled Release, 2005, 102, 247 CrossRef CAS.
- M. Lewin, N. Carlesso, C. H. Tung, X. W. Tang, D. Cory, D. T. Scadden and R. Weissleder, Nat. Biotechnol., 2000, 18, 410 CrossRef CAS.
- S. C. De Smedt, J. Demeester and W. E. Hennink, Pharm. Res., 2000, 17, 113 CrossRef.
- R. I. Mahato, A. Rolland and E. Tomlinson, Pharm. Res., 1997, 14, 853 CrossRef CAS.
- M. C. P. de Lima, S. Simoes, P. Pires, H. Faneca and N. Duzgunes, Adv. Drug Delivery Rev., 2001, 47, 277 CrossRef CAS.
- S. B. Zhang, Y. M. Xu, B. Wang, W. H. Qiao, D. L. Liu and Z. S. Li, J. Controlled Release, 2004, 100, 165 CrossRef CAS.
- B. Alberts, D. Bray, J. Lewis, M. Raff, K. Roberts and J. D. Watson, Molecular Biology of the Cell, Garland Publishing, New York, 1994 Search PubMed.
- S. M. Fuchs and R. T. Raines, Biochemistry, 2004, 43, 2438 CrossRef CAS.
- M. Belting, S. Persson and L. A. Fransson, Biochem. J., 1999, 338, 317 CrossRef CAS.
- M. Belting, Trends Biochem. Sci., 2003, 28, 145 CrossRef CAS.
- S. Sandgren, F. Cheng and M. Belting, J. Biol. Chem., 2002, 277, 38877 CrossRef CAS.
- S. Sandgren, A. Wittrup, F. Cheng, M. Jonsson, E. Eklund, S. Busch and M. Belting, J. Biol. Chem., 2004, 279, 17951 CrossRef CAS.
- K. A. Mislick and J. D. Baldeschwieler, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 12349 CrossRef CAS.
- L. C. Mounkes, W. Zhong, G. Cipres-Palacin, T. D. Heath and R. J. Debs, J. Biol. Chem., 1998, 273, 26164 CrossRef CAS.
- I. Kopatz, J. S. Remy and J. P. Behr, J. Gene Med., 2004, 6, 769 CrossRef CAS.
- C. K. Payne, S. A. Jones, C. Chen and X. W. Zhuang, Traffic, 2007, 8, 389 CrossRef CAS.
- D. L. Rabenstein, Nat. Prod. Rep., 2002, 19, 312 RSC.
- I. Lynch, T. Cedervall, M. Lundqvist, C. Cabaleiro-Lago, S. Linse and K. A. Dawson, Adv. Colloid Interface Sci., 2007, 134–135, 167 CrossRef CAS.
- A. Verma and F. Stellacci, Small, 2010, 6, 12 CrossRef CAS.
- N. L. Anderson and A. G. Anderson, Mol. Cell. Proteomics, 2002, 1, 845 CrossRef CAS.
- A. M. Alkilany, P. K. Nagaria, C. R. Hexel, T. J. Shaw, C. J. Murphy and M. D. Wyatt, Small, 2009, 5, 701 CrossRef CAS.
- K. Rezwan, L. P. Meier, M. Rezwan, J. Voros, M. Textor and L. J. Gauckler, Langmuir, 2004, 20, 10055 CrossRef CAS.
- D. Walczyk, F. B. Bombelli, M. P. Monopoli, I. Lynch and K. A. Dawson, J. Am. Chem. Soc., 2010, 132, 5761 CrossRef CAS.
- T. Xia, M. Kovochich, J. Brant, M. Hotze, J. Sempf, T. Oberley, C. Sioutas, J. I. Yeh, M. R. Wiesner and A. E. Nel, Nano Lett., 2006, 6, 1794 CrossRef CAS.
- T. Cedervall, I. Lynch, S. Lindman, T. Berggard, E. Thulin, H. Nilsson, K. A. Dawson and S. Linse, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 2050 CrossRef CAS.
- T. Cedervall, I. Lynch, M. Foy, T. Berggad, S. C. Donnelly, G. Cagney, S. Linse and K. A. Dawson, Angew. Chem., Int. Ed., 2007, 46, 5754 CrossRef CAS.
- P. McLendon, D. Buckwalter, E. Davis and T. M. Reineke, Mol. Pharmaceutics, 2010 DOI:10.1021/mp100135n.
- J. E. Schnitzer and J. Bravo, J. Biol. Chem., 1993, 268, 7562 CAS.
- J. E. Schnitzer and P. Oh, J. Biol. Chem., 1994, 269, 6072 CAS.
- J. E. Schnitzer, A. Sung, R. Horvat and J. Bravo, J. Biol. Chem., 1992, 267, 24544 CAS.
Footnotes |
| † This article is part of the ‘Emerging Investigators’ themed issue for ChemComm. |
| ‡ Electronic supplementary information (ESI) available: Detailed experimental methods and supplementary figures. See DOI: 10.1039/c0cc02618b |
| This journal is © The Royal Society of Chemistry 2011 |
