An engineered riboswitch as a potential gene-regulatory platform for reducing antibacterial drug resistance†‡
Xuli
Feng
,
Libing
Liu
*,
Xinrui
Duan
and
Shu
Wang
*
Beijing National Laboratory for Molecular Science, Key Laboratory of Organic Solids, Institute of Chemistry, Chinese Academy of Sciences, Beijing, 100190, P. R. China. E-mail: wangshu@iccas.ac.cn; liulibing@iccas.ac.cn; Fax: +86 10 6263 6680; Tel: +86 10 6263 6680
First published on 30th June 2010
Abstract
Synthetic riboswitch containing aptamer is constructed in E. coli to regulate the expression of β-lactamase through small molecule–aptamer interactions, which sharply reduces the antibiotic resistance of the engineered bacteria.
Antibiotic resistance, the ability of a microorganism to withstand the effects of antibiotics, is a growing and increasing global public health problem. The intrinsic mutation and horizontal gene transfer among bacteria have been appreciated as important forces in antibiotic resistance.1,2 The widespread applications of successive generations of antibiotics, such as β-lactams including penicillin, ampicillin, amoxicillin and cephalosporin, have made the antibiotic resistance more and more serious.3,4 Among these known mechanisms of resistance to the β-lactam antibiotics, the expression of β-lactamase is regarded as the primary one, where the antibiotic is inactivated via the hydrolysis of β-lactam ring by β-lactamase.4,5 Though great efforts have been devoted to discover new antibiotics and β-lactamase inhibitors, these strategies have failed to keep pace with the evolution of drug resistance.6,7 As a new type of antibacterial tool, antisense agents, such as RNA, PNA and DNAzymes, have been developed in bacterial systems to battle against drug resistance by inhibiting target genes.8–10 Though the principle of antisense technology is simple, it is hard to achieve regulable gene expression and uncertain toxicity is exhibited due to non-specific inhibition of other genes. Thus, new strategies to combat antibiotic resistance are still required.11
Recently, riboswitches based on small molecule–RNA interaction have attracted more and more attention due to their wide applications in controllable gene expression.12–15 Here, we construct a synthetic riboswitch containing theophylline-specific aptamer, which can regulate the genetically modified expression of β-lactamase in response to theophylline. The theophylline can induce the conformation change of the riboswitch to activate the ribozyme. The cleavage of active site results in the decrease of β-lactamase gene expression by its mRNA degradation, which ultimately leads to the diminished expression of β-lactamases. We further demonstrate that through reducing the amount of β-lactamase, the antibiotic resistance can ultimately be reduced. The small molecule theophylline can be used as co-agent to preserve the efficacy of traditional antibiotics, which provides a potential strategy to reduce antibiotic resistance instead of discovering new antibiotics.
Resistance to β-lactam antibiotic agents in Gram-negative E. coli is primarily mediated by β-lactamase, which hydrolyzes the β-lactam ring and thus inactivates the antibiotic (Scheme 1a).16 Our goal is to construct a riboswitch in the E. coli for regulating the expression of β-lactamase and reducing antibiotic resistance. The riboswitch of hammerhead ribozyme was constructed in expression vector pBV220 according to reported procedures.17 As shown in Scheme 1b, the theophylline-specific DNA aptamer sequence was inserted at a downstream location of ampicillin-resistant (Ampr) gene in the plasmid pBV220 to get new plasmid pBV220-Apt that was confirmed by restriction enzyme digestions and DNA sequencing. When integrated into the 3′-untranslated region (3′-UTR) of a reporter gene, the hammerhead ribozyme has been shown to suppress protein translation in a theophylline-dependent fashion in Saccharomyces cerevisiae.17
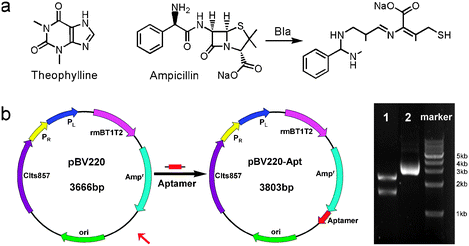 | ||
| Scheme 1 (a) Chemical structures of ampicillin and theophylline, and the hydrolytic reaction of ampicillin mediated by β-lactamase. (b) Construction map of pBV220-Apt containing the aptamer sequence and its electrophoretic analysis (Line 2) and restriction enzymes (AvrII and XhoI) cutting (Line 1). | ||
The mechanism of the RNA-based gene regulatory platform is shown in Scheme 2. In the absence of theophylline, the hammerhead ribozyme is inactive and the β-lactamase is highly expressed. Upon adding ampicillin, even with a large amount, the growth of E. coli is unaffected (Situation A). In the presence of theophylline, the ribozyme is activated through the conformational change of the theophylline-specific aptamer, and the cleavage of active site results in the decrease of β-lactamase gene expression by its mRNA degradation. Thus, the β-lactamase expression is greatly inhibited and the antibiotic resistance is sharply reduced. In this case, upon adding ampicillin, E. colicells are efficiently killed (Situation B). Therefore, the growth of bacteria cells can be effectively controlled by theophylline in the presence of ampicillin where the theophylline acts as co-agent to preserve the efficacy of traditional antibiotics.
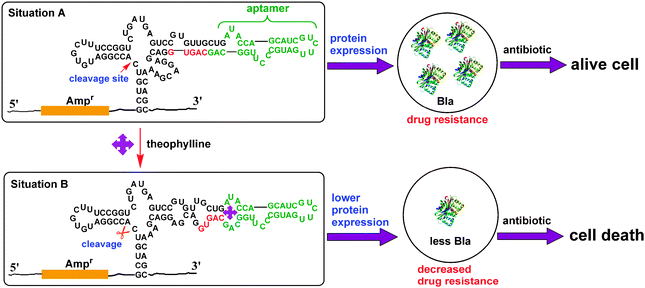 | ||
| Scheme 2 Design strategy of the RNA-based gene regulatory platform for β-lactamase expression and antibiotic resistance. The green bases indicate the sequence of the theophylline-specific aptamer. The red bases indicate the sequence of the competing strand that is part of the ribozyme. After theophylline binding to the aptamer, the secondary structure of the ribozyme is changed and the ribozyme is activated. The cleavage of active site results in the decrease of β-lactamase gene expression by its mRNA degradation, which leads to the diminished expression of β-lactamases. In this case, upon adding antibiotic, E. colicells are efficiently killed. | ||
In order to achieve the theophylline-induced gene regulation, we first tested the effect of theophylline itself on the growth of E. coli. The growth curves of E. coliTOP10 cells (Fig. 1) show that at a high concentration of theophylline (5.0 mM), the growth of TOP10 cells is inhibited. However, little toxicity is shown to the growth of E. coli for a theophylline concentration of less than 1 mM. Therefore, to ensure our gene regulatory system is strictly based on the theophylline–aptamer interactions, we carried out our following experiments at theophylline concentrations less than 1.0 mM.
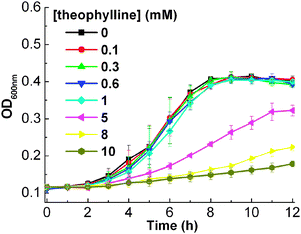 | ||
| Fig. 1 Growth curves of E. coli in the presence of varying concentrations of theophylline. The error bars represent the standard deviation of three measurements. | ||
To confirm whether the riboswitch regulates the gene expression of β-lactamase in a theophylline-dependent manner, E. coliTOP10 cells transformed plasmid pBV220 or ribozyme modified pBV220-Apt were grown in LB media in the presence of ampicillin (50 μg mL−1). The β-lactamase activity was measured using a spectrophotometric assay with nitrocefin as the chromogenic cephalosporin substrate18 with varying concentrations of theophylline from 0 to 1 mM (Fig. 2). The addition of theophylline produces a significantly dose-dependent decrease in β-lactamase activity of E. coliTOP10 cells transformed plasmid pBV220-Apt, whereas little change of β-lactamase activity was observed for the E. coliTOP10 cells without the riboswitch. These results imply that the expression of β-lactamase can be effectively suppressed by non-antibiotic theophylline, thus providing a controllable way to modulate the antibiotic resistance to the engineered bacteria.
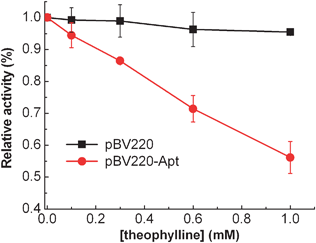 | ||
| Fig. 2 Riboswitch mediated β-lactamase relative activity in E. coliTOP10 cells as a function of varying concentrations of theophylline from 0 to 1.0 mM. The error bars represent the standard deviation of three measurements. | ||
In order to reduce drug resistance and inhibit the growth of bacteria, E. coli harboring pBV220 or pBV220-Apt were grown on LB/agar supplemented with ampicillin (3.0 mg mL−1). Fig. 3a shows the results of plate-based growth of TOP10 cells that were grown at 37 °C for 18 h. It's clear that with the increasing concentration of ampicillin up to 3.0 mg mL−1, cells transformed plasmid pBV220 can still grow well, which indicates the host cells have stronger antibiotic resistance. After adding 1.0 mM theophylline, the growth of the cells was unaffected, which guarantees that the theophylline itself has no effect on the growth of E. coli under this condition. For the host cells with riboswitch, without theophylline, they can grow well even in the presence of a high concentration of ampicillin (3.0 mg mL−1). However, in the presence of 1.0 mM theophylline, the riboswitch in host cells was activated, leading to reduced expression of β-lactamase. Thus, the antibiotic resistance can be diminished and these cells can hardly grow under this circumstance. Since the gene regulatory system is based on theophylline–aptamer interactions, the antibiotic resistance of E. coli harboring plasmid pBV220-Apt can be controlled in a theophylline concentration-dependent manner. As shown in Fig. 3b, in the presence of ampicillin (3.0 mg mL−1), the growth of TOP10 cells can be effectively inhibited by increasing the concentrations of theophylline. Therefore, the small molecule theophylline can be used as co-agent to preserve the efficacy of traditional antibiotics.
![(a) The colony forming units (CFU) for E. coli on LB agar plate for pBV220 and riboswitchable pBV220-Apt in the absence or presence of 1.0 mM theophylline. [ampicillin] = 3.0 mg mL−1. (b) The percentage of colony forming units for E. coli harboring riboswitchable pBV220-3′ as a function of varying concentrations of theophylline. [ampicillin] = 3.0 mg mL−1. [theophylline] = 0–1.0 mM. The error bars represent the standard deviation of three measurements.](/image/article/2011/CC/c0cc00980f/c0cc00980f-f3.gif) | ||
| Fig. 3 (a) The colony forming units (CFU) for E. coli on LB agar plate for pBV220 and riboswitchable pBV220-Apt in the absence or presence of 1.0 mM theophylline. [ampicillin] = 3.0 mg mL−1. (b) The percentage of colony forming units for E. coli harboring riboswitchable pBV220-3′ as a function of varying concentrations of theophylline. [ampicillin] = 3.0 mg mL−1. [theophylline] = 0–1.0 mM. The error bars represent the standard deviation of three measurements. | ||
In summary, we demonstrate that a synthetic riboswitch containing a theophylline-specific aptamer sequence can regulate the expression of β-lactamase through theophylline–aptamer interactions, which sharply reduces the antibiotic resistance of the engineered bacteria. The non-antibiotic theophylline can be used as co-agent to preserve the efficacy of traditional antibiotics. Thus a non-antibiotic small molecule induced RNA gene regulatory system could provide an alternative and potential way to control drug resistance. This new strategy also provides a useful tool for understanding the antibiotic resistance-related bioprocess and gene regulation. Since gene transfer happens among bacterial cells, the leakage of engineered genes can be potentially applied to control the antibiotic resistance of non-engineered pathogenic bacteria. Of course, there is a lot of work to do before the strategy can be used in non-engineered bacteria.
The authors are grateful to the National Natural Science Foundation of China (Nos. 20725308, 90913014, 20721061 and TRR61) and the Major Research Plan of China (No. 2006CB932102).
Notes and references
- M. Lipsitch and M. H. Samore, Emerging Infectious Diseases, 2002, 8, 347–354 Search PubMed.
- J. L. Martínez, F. Baquero and D. I. Andersson, Nat. Rev. Microbiol., 2007, 5, 958–965 Search PubMed.
- J. L. Davies, Science, 1994, 264, 375–382 CrossRef CAS.
- C. Walsh, Nature, 2000, 406, 775–781 CrossRef CAS.
- J. F. Fisher, S. O. Meroueh and S. Mobashery, Chem. Rev., 2005, 105, 395–424 CrossRef CAS.
- L. Miesel, J. Greene and T. A. Black, Nat. Rev. Genet., 2003, 4, 442–456 CrossRef CAS.
- D. Hughes, Nat. Rev. Genet., 2003, 4, 432–441 CrossRef CAS.
- A. Nikravesh, R. Dryselius, O. R. Faridani, S. Goh, M. Sadeghizadeh, M. Behmanesh, A. Ganyu, E. J. Klok, R. Zain and L. Good, Mol. Ther., 2007, 15, 1537–1542 CrossRef CAS.
- Y. D. Ji, B. Zhang, S. F. Van Horn, P. Warren, G. Woodnutt, M. K. R. Burnham and M. Rosenberg, Science, 2001, 293, 2266–2269 CrossRef CAS.
- L. Good, S. K. Awasthi, R. Dryselius, O. Larsson and P. E. Nielsen, Nat. Biotechnol., 2001, 19, 360–364 CrossRef CAS.
- P. A. Smith and F. E. Romesberg, Nat. Chem. Biol., 2007, 3, 549–556 CrossRef CAS.
- W. C. Winkler, A. Nahvi, A. Roth, J. A. Collins and R. R. Breaker, Nature, 2004, 428, 281–286 CrossRef CAS.
- V. Sharma, Y. Nomura and Y. Yokobayashi, J. Am. Chem. Soc., 2008, 130, 16310–16315 CrossRef CAS.
- T. S. Bayer and C. D. Smolke, Nat. Biotechnol., 2005, 23, 337–343 CrossRef CAS.
- S. K. Desai and J. P. Gallivan, J. Am. Chem. Soc., 2004, 126, 13247–13254 CrossRef CAS.
- L. Brinas, M. Zarazaga, Y. Saenz, F. Ruiz-Larrea and C. Torres, Antimicrob. Agents Chemother., 2002, 46, 3156–3163 CrossRef CAS.
- M. N. Win and C. D. Smolke, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 14283–14288 CrossRef CAS.
- M. Hedberg, L. Lindqvist, T. Bergman and C. E. Nord, Antimicrob. Agents Chemother., 1995, 39, 1458–1461 CAS.
Footnotes |
| † This article is part of the ‘Emerging Investigators’ themed issue for ChemComm. |
| ‡ Electronic supplementary information (ESI) available: Details for the experiments and measurements. DNA sequencing of synthetic ribozyme. See DOI: 10.1039/c0cc00980f |
| This journal is © The Royal Society of Chemistry 2011 |
