Synthesis and application of a new cleavable linker for “click”-based affinity chromatography†‡
Felicetta Landi, Conny M. Johansson, Dominic J. Campopiano and Alison N. Hulme*
School of Chemistry, The University of Edinburgh, Kings Buildings, West Mains Road, Edinburgh, UK EH9 3JJ. E-mail: Alison.Hulme@ed.ac.uk; Fax: +44 131 650 4743; Tel: +44 131 650 4711
First published on 13th October 2009
Abstract
A new chemically-cleavable linker has been synthesised for the affinity-independent elution of biomolecules by classical affinity chromatography. This azo-based linker is shown to couple efficiently with “click” derivatised ligands such as biotin propargyl amide through a copper(I)-catalysed Huisgen 1,3-dipolar cycloaddition reaction. Binding to Affi-Gel matrices displaying ligands coupled to the new linker is both efficient and selective. The captured material may be readily released from the resin upon treatment with sodium dithionite. These mild elution conditions have allowed for the efficient isolation of the affinity partner from complex protein mixtures such as those found in fetal bovine serum.
Introduction
Affinity chromatography has relied historically on specific bioaffinity interactions between a resin-bound ligand and its biomolecular partner to effect the separation of that partner from a complex mixture of components (Fig. 1).1 For successful application, the coupled ligand must retain its specific binding affinity for the target molecule and, once the unbound material has been removed by washing, the binding between the ligand and the target molecule must be reversible to allow the target molecule to be removed in an active form.2 Perhaps one of the most notable early examples of the use of affinity chromatography for the identification of the binding partner of small-molecules was the capture of the rapamycin- and FK506-binding protein (FKBP) and other immunophilins from a human Jurkat T cell line extract.3 More recently, affinity chromatography has been applied to the field of proteomics either in combination with small molecule probes for activity-based protein profiling to generate specific sub-proteomes,4 or through amino acid-specific labelling of proteins (e.g. with biotin). This can be done either prior to, or post, trypsin digest to allow pre-fractionation of a complex mixture of peptides through an intermediate purification protocol (e.g. on streptavidin-coated beads) before MS analysis.5,6 However, the efficient recovery of the separated biomaterial in a functionally active form can be a difficult process since it often requires elution using high salt concentrations and/or detergents [Fig. 1(a)].2 For this reason, general elution methods which are independent of the bioaffinity interaction (so-called affinity-independent elution protocols) are of particular interest.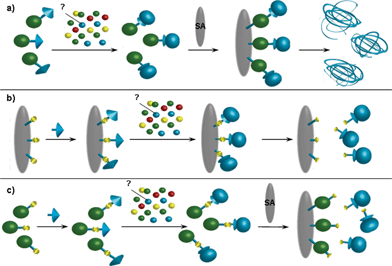 | ||
| Fig. 1 Capture of an “unknown” affinity target for a small-molecule ligand: (a) capture of the affinity target in solution with a biotinylated ligand, immobilisation of the biotinylated ligand complex on a (strept)avidin coated matrix and elution under potentially denaturing conditions; (b) direct attachment of a “click” functionalised ligand to the matrix through a cleavable linker, capture of the affinity target and elution under affinity-independent, non-denaturing conditions; (c) attachment of biotin to the ligand through a cleavable linker, capture of the affinity target in solution, immobilisation of the biotinylated ligand complex on a (strept)avidin coated matrix and elution under affinity-independent, non-denaturing conditions. | ||
Affinity-independent elution of the captured biomaterial [Fig. 1(b) & (c)] typically relies upon the introduction of a linker between the resin and the bound ligand. This linker may be enzymatically cleaved (e.g. the nuclease-cleavable linker described by Santala et al. for the efficient recovery of antibody-displaying phages that have been captured with a biotinylated antigen on streptavidin-coated magnetic particles),7 photocleavable (e.g. the photocleavable linker applied to microbead-based affinity chromatography chips used to purify the hepatitis C virus RNA polymerase proteins described by Cho et al.),8 or cleaved under mild chemical conditions (e.g. the use of an azobenzene linker to purify cathepsin peptides from rat liver homogenate described by Verhelst et al.).9a This last case was of particular interest to us as the cleavage reaction conditions (mild reduction with sodium dithionite, Na2S2O4) are known to be compatible with biochemical systems,§ as exemplified by the use of azobenzenes as cleavable cross-linking reagents for proteins10 and in the functionalisation of a tyrosine residue on the surface of viral capsids.11 Thus in designing a cleavable support for affinity-independent elution methodology that was compatible with the extensive range of propargyl functionalised “click” ligands that have emerged in recent years,12,13 we chose to base our linker on this precedent.
Results and discussion
Azobenzene linker 1 was designed with orthogonal reactive functionalities: an amine for linking to a matrix support, and an azide for reaction with the propargyl-functionalised ligands of interest. The azobenzene unit (2) at the heart of this linker was found to be readily accessible through diazotisation of 4-aminobenzoic acid and reaction with phenol (Scheme 1).14 Etherification of the phenol in the presence of the benzoic acid was found to give a mixture of products, but reaction of the acid with N-Boc-ethylenediamine15 followed by phenol etherification with 1,3-dibromopropane16 resulted in a good yield of alkyl bromide 3. Bromide displacement (NaN3, 3→4) and Boc-deprotection, led to the preparation of a novel cleavable linker 1 for affinity applications which could be readily prepared on a gram scale (5 steps, 29% overall yield).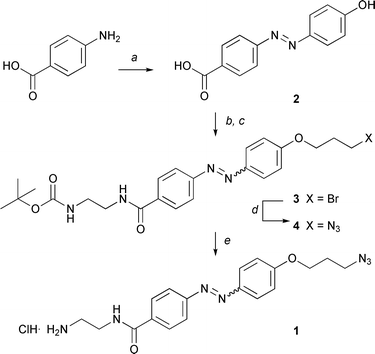 | ||
| Scheme 1 Linker synthesis. Reagents and conditions: (a) i. NaNO2, HCl, 0 °C, 20 min; ii. PhOH, NaOH, K2CO3, 0 °C to RT, 3 h, 90%; (b) BocNHCH2CH2NH2, EDC, HOBt, DMF, 18 h, 60%; (c) 1,3-dibromo-propane, K2CO3, CH3CN reflux, 5 h, 67%; (d) NaN3, DMF, 18 h, 90%; (e) MeOH, CH3COCl, 0 °C to RT, Et2O, 1 h, 90%. | ||
For our preliminary affinity chromatography studies we chose to use the matrix Affi-Gel 10 (Bio-Rad), an agarose based affinity medium that contains a neutral 10-atom spacer arm and a reactive N-hydroxysuccinimide ester (Scheme 2). Complementary supported azides 5 and 6 (with and without the azo linker) were generated through coupling either 3-azidopropylamine, or linker 1 to the Affi-Gel 10 matrix. A ligand-assisted, copper(I)-catalysed Huisgen 1,3-dipolar cycloaddition, or “click”, reaction conducted in the presence of tris-(benzyltriazolylmethyl)amine (TBTA)17 allowed successful coupling of propargyl alcohol to azide 6 (Scheme 2). This derivatised matrix 7 was used to test the desired chemoselective reductive cleavage of the azo linker by sodium dithionite (Na2S2O4)18 which was easily followed since reduction of the azo group is accompanied by a dramatic loss of colour of the agarose matrix. The corresponding aniline derivative 8 was eluted from matrix 7 in good yield (70%). This preliminary study demonstrates for the first time that the azo unit is compatible with the reductive conditions of the “click” coupling reaction.
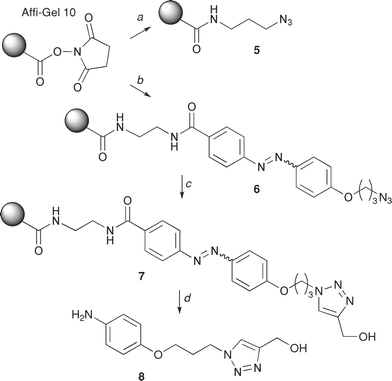 | ||
| Scheme 2 Affi-Gel supported azide synthesis and reactions. Reagents and conditions: (a) 3-azidopropylamine, MeOH, 18 h; (b) linker 1, Et3N, H2O/CH3CN, 18 h; (c) propargyl alcohol, CuSO4, sodium ascorbate, TBTA, H2O, t-BuOH, 18 h; (d) Na2S2O4. | ||
In order to test how robust the new linker would be in an affinity-independent elution protocol, the biotin/(strept)avidin affinity pair was selected. Biotin and its affinity partner (strept)avidin exhibit one of the strongest known non-covalent interactions with a dissociation constant of KD≈ 10−15 M and they are widely used in affinity based separations.19 Although direct attachment of biotin to the matrix and isolation of its known binding partner, avidin, was chosen to investigate the new click linker properties (Fig. 1b, “unknown” target = avidin), this study would also probe the ability of the linker to provide an affinity-independent elution protocol where a click functionalised ligand was coupled to a biotinylated derivative of the linker and a (strept)avidin-coated matrix was used to isolate the unknown target of the ligand following incubation (Fig. 1c).¶ Indeed, the aqueous-based “click” coupling reaction of biotin-linker conjugates has been shown to have many advantages over conventional coupling techniques as a means of selective ligand biotinylation.20
The propargyl amide derivative of biotin21 was coupled directly to the azido matrix 5 to generate affinity matrix 9, as well as through the cleavable azobenzene unit in 6 to generate affinity matrix 10 (Scheme 3). In the first set of affinity experiments designed to test the new linker for its ability to bind the affinity partner without any competing non-specific interactions, both matrices 9 and 10 were incubated with avidin, whilst only matrix 10 with its novel azo “click” linker was incubated with albumin (negative control). The absence of a clear band for avidin (15 kDa) in the supernatant following incubation shows that both matrices bound avidin (Fig. 2, lanes A, B, D & E). Furthermore, matrix 10 was demonstrated not to exhibit any non-specific interactions for albumin as shown by its quantitative elution; clear band at 66 kDa (Fig. 2, lane C) and the absence of this band following cleavage (Fig. 2, lane G).
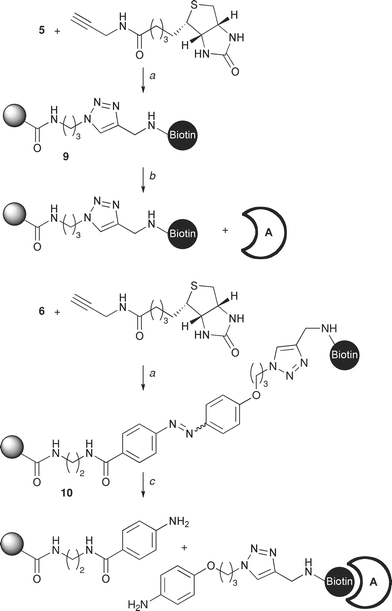 | ||
| Scheme 3 Preliminary affinity studies with derivatised Affi-Gel matrices 5 and 6. Reagents and conditions: (a) biotin propargyl amide, CuSO4, sodium ascorbate, TBTA, H2O, t-BuOH, 18 h; (b) incubation with avidin followed by elution with SDS; (c) incubation with avidin followed by cleavage with Na2S2O4. | ||
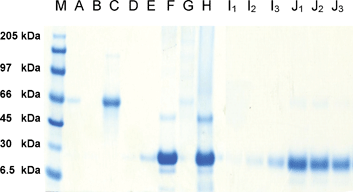 | ||
| Fig. 2 SDS-PAGE for experiments with avidin and albumin: lanes A-E supernatants after incubation; lanes F-H supernatants following SDS elution; lanes I1-J3 supernatants following dithionite cleavage. M = marker lane; A & B = supernatant from incubation of matrix 9 with avidin; C = supernatant from incubation of matrix 10 with albumin; D & E = supernatant from incubation of matrix 10 with avidin; F = supernatant after SDS elution of matrix 9 incubated with avidin (Lane A); G = supernatant after SDS elution of matrix 10 incubated with albumin (Lane C); H = supernatant after SDS elution of matrix 10 incubated with avidin (Lane D); I1-I3 = supernatant after dithionite cleavage of matrix 9 incubated with avidin (Lane B) at pH 6.5, 7.5 & 8.5; J1-J3 = supernatant after dithionite cleavage of matrix 10 incubated with avidin (Lane E) at pH 6.5, 7.5 & 8.5. | ||
The efficiency of a classical SDS elution procedure (for matrix 9) and chemoselective cleavage with sodium dithionite (for matrix 10) were then compared. Gratifyingly, both elution procedures showed the expected bands for avidin (Fig. 2, lanes F & J1-3). The efficiency of dithionite cleavage was determined at a range of pH values (Fig. 2, lanes J1-3); incubation with a phosphate buffer solution at pH 6.5 and a large excess of sodium dithionite were identified as optimal cleavage conditions (Fig. 2, lane J1). These mild cleavage conditions were shown not to release avidin from matrix 9 in the absence of the azo linker (Fig. 2, lanes I1-3) demonstrating the chemoselectivity of this protocol. Conversely, the presence of the linker does not interfere with a classical SDS elution protocol, as avidin is released from matrix 10 under these conditions (Fig. 2, lane H).
In the second set of experiments, matrices 9 and 10 were incubated with avidin enriched fetal bovine serum (FBS) (Fig. 3, lane A) to determine whether the derivatised Affi-Gel matrices could capture avidin selectively from complex protein mixtures. Gratifyingly, both matrices were shown to remove avidin (Fig. 3, lanes B & C). Furthermore, release of the bound material could be performed using the optimal cleavage conditions for each matrix: boiling with SDS for matrix 9 (Fig. 3, lane D); and dithionite cleavage at pH 6.5 for matrix 10 (Fig. 3, lane E),|| with only marginally reduced efficiency for the chemical cleavage.
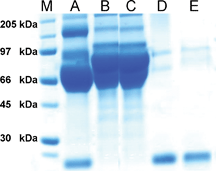 | ||
| Fig. 3 SDS-PAGE for experiments with FBS: M = marker lane; A = FBS + avidin (control lane); B = supernatant after incubation of matrix 9 with FBS + avidin; C = supernatant after incubation of matrix 10 with FBS + avidin; D = supernatant after SDS elution of matrix 9 incubated with FBS + avidin (Lane B); E = supernatant after dithionite cleavage of matrix 10 incubated with FBS + avidin (Lane C). | ||
Conclusion
A new azobenzene-based linker for affinity chromatography applications has been developed; it may be readily cleaved under mild non-denaturing conditions (elution with Na2S2O4) making it suitable for affinity-independent elution protocols. Affinity ligands functionalised with a bioorthogonal propargyl label may be readily attached to the terminal azide of the linker using the copper(I)-catalysed Huisgen 1,3-dipolar cycloaddition reaction (or “click” reaction). Affinity capture by matrices displaying ligands coupled to the new linker has been shown to be both efficient and selective. The chemical cleavage of the ligand plus its bioaffinity partner from the gel has been shown to be comparable to a classical cleavage process under denaturing conditions; and indeed the two processes are both compatible with the new linker. Although the use of this linker has been demonstrated using the agarose matrix Affi-Gel 10, we believe that it will find widespread application on a range of activated matrix and/or chip surfaces. It is anticipated that the use of this azo linker as means of coupling biotin to the ligand of interest will also allow an affinity-independent version of the more classical manifold for target identification based upon immobilisation on a (strept)avidin coated matrix, as well as allowing in vivo experiments. Investigations with several ligands of interest (whose propargyl derivatives retain the activity of the lead compound) with known, or potential, affinity partners are currently under progress in our research group.Acknowledgements
We thank the EC FP6 for funding (Marie Curie EST Fellowships to FL and CMJ; Contract: MEST-CT-2005-020744) and Gareth Morrison for helpful discussions about the preliminary affinity studies.Notes and references
- P. Cuatrecasas and C. B. Anfinsen, Annu. Rev. Biochem., 1971, 40, 259–278 CrossRef CAS; Affinity Chromatography–Principles and Methods, Amersham Bioscience, 2003 Search PubMed.
- M. A. Firer, J. Biochem. Biophys. Methods, 2001, 49, 433–442 CrossRef CAS.
- H. Fretz, M. W. Albers, A. Galat, R. F. Standaert, W. S. Lane, S. J. Burakoff, B. E. Bierer and S. L. Schreiber, J. Am. Chem. Soc., 1991, 113, 1409–1411 CrossRef CAS.
- H. Ovaa and F. van Leeuwen, ChemBioChem, 2008, 9, 2913–2919 CrossRef CAS; S. A. Sieber and B. F. Cravatt, Chem. Commun., 2006, 2311–2319 RSC.
- A. Leitner and W. Lindner, J. Chromatogr., B, 2004, 813, 1–26 CrossRef CAS.
- R. Aebersold and M. Mann, Nature, 2003, 422, 198–207 CrossRef CAS.
- V. Santala and P. Saviranta, J. Immunol. Methods, 2004, 284, 159–163 CrossRef CAS.
- S. Cho, S.-H. Lee, W.-J. Chung, Y.-K. Kim, Y.-S. Lee and B.-G. Kim, Electrophoresis, 2004, 25, 3730–3739 CrossRef CAS.
- (a) S. H. L. Verhelst, M. Fonovic and M. Bogyo, Angew. Chem., Int. Ed., 2007, 46, 1284–1286 CrossRef CAS; (b) N. Abello, H. A. M. Kerstjens, D. S. Postma and R. Bischoff, J. Proteome Res., 2009, 8, 3222–3238 CrossRef CAS.
- H. R. Murphy and H. W. Harris, Jr., Anal. Biochem., 1987, 165, 88–95 CrossRef CAS.
- J. M. Hooker, E. W. Kovacs and M. B. Francis, J. Am. Chem. Soc., 2004, 126, 3718–3719 CrossRef CAS.
- M. D. Best, Biochemistry, 2009, 48, 6571–6584 CrossRef CAS; J. E. Moses and A. D. Moorhouse, Chem. Soc. Rev., 2007, 36, 1249–1262 RSC.
- The complementary strategy of azide-functionalised “click” ligand capture has been reported: M. A. Nessen, G. Kramer, J. W. Back, J. M. Baskin, L. E. J. Smeenk, L. J. de Koning, J. H. van Maarseveen, L. de Jong, C. R. Bertozzi, H. Hiemstra and C. G. de Koster, J. Proteome Res., 2009, 8, 3702–3711 Search PubMed; J. Gubbens, E. Ruijter, L. E. V. de Fays, J. M. A. Damen, B. de Kruijff, M. Slijper, D. T. S. Rijkers, R. M. J. Liskamp and A. I. P. M. de Kroon, Chem. Biol., 2009, 16, 3–14 CrossRef CAS.
- R. J. Cooper, P. J. Camp, R. J. Gordon, D. K. Henderson, D. C. R. Henry, H. McNab, S. S. de Silva, D. Tackley, P. A. Tasker and P. Wight, Dalton Trans., 2006, 2785–2793 RSC; Y. Yu, T. Maeda, J.-I. Mamiya and T. Ikeda, Angew. Chem., Int. Ed., 2007, 46, 881–883 CrossRef CAS.
- J. Ohkanda and A. Katoh, Tetrahedron, 1995, 51, 12995–13002 CrossRef CAS.
- J. T. Strupczewski, K. J. Bordeau, Y. Chiang, E. J. Glamkowski, P. G. Conway, R. Corbett, H. B. Hartman, M. R. Szewczak, C. A. Wilmot and G. C. Helsley, J. Med. Chem., 1995, 38, 1119–1131 CrossRef CAS.
- T. R. Chan, R. Hilgraf, K. B. Sharpless and V. V. Fokin, Org. Lett., 2004, 6, 2853–2855 CrossRef CAS.
- R. D. Voyksner, R. Straub and J. T. Keever, Environ. Sci. Technol., 1993, 27, 1665–1672 CAS; L.-H. Ahlström, C. S. Eskilsson and E. Björklund, Trends Anal. Chem., 2005, 24, 49–56 CrossRef CAS.
- E. A. Bayer and M. Wilchek, J. Chromatogr., A, 1990, 510, 3–11 CrossRef CAS.
- I. A. Inverarity, R. F. H. Viguier, P. Cohen and A. N. Hulme, Bioconjugate Chem., 2007, 18, 1593–1603 CrossRef CAS; S. Peddibhotla, Y. Dang, J. O. Liu and D. Romo, J. Am. Chem. Soc., 2007, 129, 12222–12231 CrossRef.
- X. Z. Zhao, E. A. Semenova, C. Liao, M. Nicklaus, Y. Pommier and T. R. Burke, Jr., Bioorg. Med. Chem., 2006, 14, 7816–7825 CrossRef CAS.
Footnotes |
| † This paper is part of an Organic & Biomolecular Chemistry web theme issue on protein labelling and chemical proteomics. Guest editors: Ed Tate and Matthew Bogyo. |
| ‡ Electronic supplementary information (ESI) available: Details of the synthesis of linker 1, preparation and affinity applications of derivatised Affi-Gel matrices 7, 9 and 10, together with spectra for the linker and azides. See DOI: 10.1039/b916693a |
| § Whilst this reduction has found widespread use in biochemical applications, the concomitant reduction of some amino acids such as nitrotyrosine to aminotyrosine has been reported under these conditions.9b |
| ¶ Synthetically this would require coupling of the amino group of the azo linker 1 to NHS-biotin, followed by click coupling of the biotin-linker conjugate to the ligand. |
| || The weak bands at 66-97 kDa are likely to be serum albumin and similar prominent serum proteins. |
| This journal is © The Royal Society of Chemistry 2010 |
