 Open Access Article
Open Access ArticleGlycosidase inhibition: assessing mimicry of the transition state
Tracey M. Gloster*ab and Gideon J. Davies*a
aYork Structural Biology Laboratory, Department of Chemistry, University of York, Heslington, York YO10 5YW, UK. E-mail: davies@ysbl.york.ac.uk; gloster@ysbl.york.ac.uk; Fax: +44 1904 328266; Tel: +44 1904 328260
bDepartment of Chemistry, Simon Fraser University, 8888 University Drive, Burnaby, BC V5A 1S6, Canada
First published on 5th November 2009
Abstract
Glycoside hydrolases, the enzymes responsible for hydrolysis of the glycosidic bond in di-, oligo- and polysaccharides, and glycoconjugates, are ubiquitous in Nature and fundamental to existence. The extreme stability of the glycosidic bond has meant these enzymes have evolved into highly proficient catalysts, with an estimated 1017 fold rate enhancement over the uncatalysed reaction. Such rate enhancements mean that enzymes bind the substrate at the transition state with extraordinary affinity; the dissociation constant for the transition state is predicted to be 10−22 M. Inhibition of glycoside hydrolases has widespread application in the treatment of viral infections, such as influenza and HIV, lysosomal storage disorders, cancer and diabetes. If inhibitors are designed to mimic the transition state, it should be possible to harness some of the transition state affinity, resulting in highly potent and specific drugs. Here we examine a number of glycosidase inhibitors which have been developed over the past half century, either by Nature or synthetically by man. A number of criteria have been proposed to ascertain which of these inhibitors are true transition state mimics, but these features have only be critically investigated in a very few cases.
 Tracey M. Gloster | Tracey graduated with a B.Sc. in Biochemistry from the University of Warwick (2002), which was followed by a Ph.D. at the University of York under the supervision of Prof. Gideon Davies (2006). This work involved the structural, kinetic and thermodynamic characterisation of a panel of transition state analogues with glycoside hydrolases. Following a short period as a post-doctoral fellow in the same laboratory, Tracey was awarded a Sir Henry Wellcome post-doctoral fellowship by the Wellcome Trust, which started in 2008. Tracey is currently working with David Vocadlo at Simon Fraser University investigating the modulation of the O-GlcNAc post-translational modification. |
 Gideon J. Davies | Gideon Davies followed his B.Sc. in Biochemistry from the University of Bristol with a Ph.D. in 1990. Gideon subsequently moved to the EMBL Hamburg Outstation to work with Keith Wilson. Later in the same year, Gideon moved to York to work with Dale Wigley and Guy Dodson on the structure of DNA gyrase, and continued in York where he started working on carbohydrate-active enzymes. In 1996 Gideon was awarded a Royal Society University Research Fellowship and made a Professor at the University of York in 2001. Gideon won the Roy L. Whistler prize of the International Carbohydrate Organization in 2006. |
Introduction
Glycosidases, the enzymes responsible for the breakdown of di-, oligo- and polysaccharides, and glyconjugates, are ubiquitous through all kingdoms of life. Carbohydrate processing enzymes, including glycosidases and glycosyltransferases (the enzymes which transfer saccharides to other saccharide moieties, small molecules, lipids or proteins), constitute between 1 and 3% of the genome of most organisms.1 The task facing these enzymes with respect to maintaining efficient and highly specific catalysis is no mean feat; it has been calculated that there are 1.05 × 1012 possible linear and branched forms of a hexasaccharide2 and that carbohydrates account for around 75% of the biomass on Earth. The extreme stability of the glycosidic bond and the catalytic rates glycosidases achieve mean they are among the most proficient of all enzymes.3 Although glycosidases and glycosyltransferases act on a huge range of differing substrates, individual enzymes must display specificity related to their function. Indeed, the roles of these enzymes are numerous and diverse ranging from glycosylation of proteins in the Golgi apparatus to plant cell wall biosynthesis, from breakdown of ingested material in the gut to defence mechanisms against microbial infection.Great efforts have been made in recent years to design and synthesize inhibitors of glycosidases. Given their multitude of roles in vivo, inhibition of these enzymes in a number of different processes is extremely attractive with potential in the treatment of lysosomal storage diseases,4–7 diabetes8,9 and viral infections10 including influenza11,12 and HIV;13 indeed there are a number of drugs currently on the market which are used in the treatment of some of these diseases. Acarbose14 and Miglitol15 target intestinal tract α-glucosidases and α-amylases in the treatment of type II diabetes. These enzymes are responsible for the digestion of dietary carbohydrates to monosaccharides that can be absorbed through the intestinal wall into the blood stream; their inhibition helps to control uptake of monosaccharides into the blood and suppress an excessive rise in blood glucose. Relenza16,17 and Tamiflu18,19 target a neuraminidase on the influenza virus surface, which during infection cleaves terminal sialic acid residues to destroy the receptors recognised by haemagglutinin, and thus plays an important role in spreading the virus and preventing aggregation of viral particles. Inhibition of the neuraminidase causes the spread of the progeny virus to new cells to be slowed or halted. As with any new drug, the key to producing clinically relevant glycosidase inhibitors is not only high potency, but also specificity over other enzymes which may be encountered. In addition to potential therapeutic applications, glycosidase inhibitors are extremely useful probes for dissecting the subtle differences in mechanism employed by different glycosidase families (see below) and also as tools for chemical biology. In such a regard selective inhibitors can be invaluable for modulating effects in cells or in vivo (for example see Ref. 20, 21).
CAZy families
Carbohydrate processing enzymes are classified by primary sequence similarity into ‘families’, which are listed in the Carbohydrate Active enZyme (CAZy) database22 (available at http://www.cazy.org); at present there are 115 sequence-distinct families of glycosidases. A feature of most CAZy families is that as the primary sequence dictates structure, and structure determines function, the catalytic mechanism is conserved within a family.23 There are, however, some exceptions such as the NAD+-dependent enzymes in GH424 and GH10925 (discussed further below), GH97 enzymes have recently been shown to contain two sub-families which act with inversion and retention of configuration,26,27 and the GH23 enzymes. Family GH23 contains goose type lysozymes, which hydrolyse with inversion of stereochemistry,28 and peptidoglycan lytic transglycosylases, which use an intramolecular rearrangement, with retention of configuration, to form an 1,6-anhydrosugar product;29 the reaction mechanisms involved, however, remain unclear.Glycosidase mechanisms
Hydrolysis of the glycosidic bond proceeds with either net retention or inversion of anomeric configuration. The ‘classical’ mechanisms for glycoside hydrolysis were first proposed by Koshland in 195330 and, now over 50 years later, have stood the test of time and a vast amount of biochemical investigation and remain largely unchanged (for reviews see Ref. 31–33). Traditionally (although there are exceptions) classical glycosidases possess two carboxylate-containing residues which are responsible for hydrolysis. Inversion of stereochemistry is a single step mechanism (Fig. 1a), which allows both substrate and a water molecule to be bound simultaneously. One of the catalytic residues acts as a general acid and the other as a general base. Protonation of the glycosidic oxygen by the general acid and departure of the leaving group is accompanied by concomitant nucleophilic attack by a water molecule that has been deprotonated by the general base.34,35 Retention of stereochemistry is a double displacement mechanism, consisting of two inverting steps (Fig. 1b); one of the catalytic residues acts as the acid/base residue and the other as a nucleophile. During the first (glycosylation) step of the reaction the acid/base protonates the glycosidic oxygen to aid leaving group departure, which is concomitant with attack of the nucleophile at the anomeric carbon, and leads to formation of a covalent glycosyl-enzyme intermediate. In the second (deglycosylation) step the acid/base residue deprotonates a water molecule which provides a nucleophilic species to attack at the anomeric carbon and displace the glycoside.34,35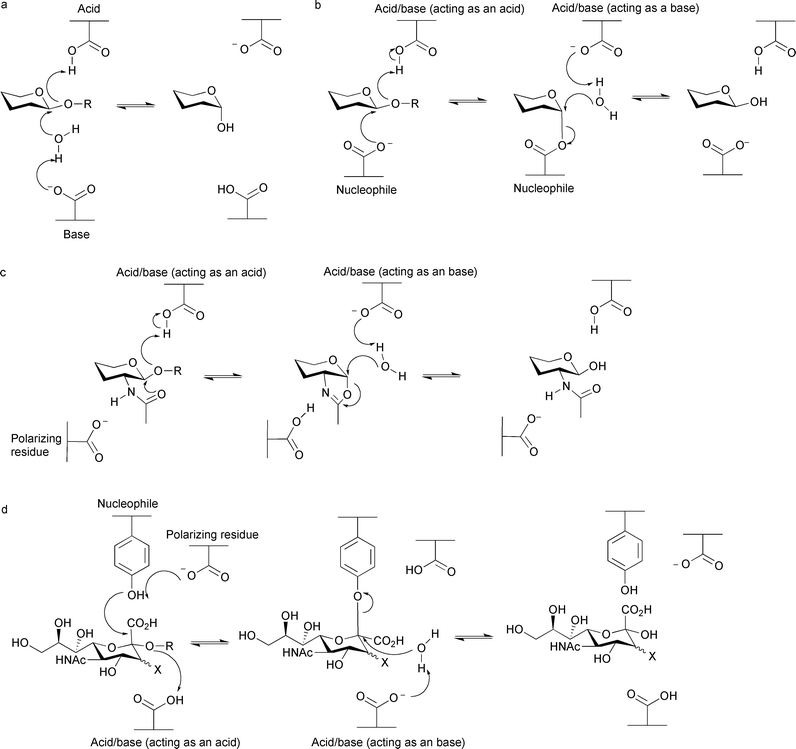 | ||
| Fig. 1 Glycosidase mechanisms for hydrolysis. (a) ‘Classical’ mechanism for inversion of stereochemistry. (b) ‘Classical’ mechanism for retention of stereochemistry. (c) Substrate-assisted mechanism proposed for families 18, 20, 56, 84, 85 and possibly 25. (d) Mechanism using a tyrosine residue as the nucleophile proposed for families 33 and 34. | ||
As structural and mechanistic studies have become more sophisticated, other mechanisms have been proposed for small subsets of glycoside hydrolases, and this information aids inhibitor design for these enzymes. A number of families containing glycosidases which hydrolyse substrates containing N-acetylhexosamine with retention of configuration (which are classified into families GH18,36 GH20,37 GH56,38 GH84,39,40 GH85,41,42 and is likely for GH2543) have been shown to lack a conventional catalytic nucleophile, but instead use a substrate-assisted catalytic mechanism (Fig. 1c). The acetamido group at the C2 position of the substrate acts as a nucleophile to attack the anomeric carbon to create an enzyme-stabilised oxazoline intermediate. The intermediate is hydrolysed by a water molecule, which is activated by a residue acting as a general base. In most cases, a second carboxylate-containing residue orients and polarizes the 2-acetamido group to increase its nucleophilicity. Some sialidases and neuraminidases (such as those classified into families GH33 and GH34) catalyse hydrolysis of sialic acid-containing substrates (in an exo fashion, where only the terminal residue is cleaved), with retention of configuration, using a tyrosine residue as the catalytic nucleophile.44,45 This differs from the ‘classical’ mechanism where traditionally a residue harbouring a carboxylate group is responsible for nucleophilic attack, but a similar mechanism is employed (Fig. 1d). The tyrosine residue relays charge from a nearby glutamate residue to provide a nucleophilic oxygen atom carrying some negative charge; it is proposed a glutamate residue in this position would cause electrostatic repulsions with the carboxylate group of the sialic acid. Other sialidases and neuraminidases (such as those classified in families GH5846 and GH8347), which hydrolyse sialic acid-containing substrates in an endo fashion (where cleavage occurs within a polysaccharide), have been observed to lack the tyrosine residue which acts as a nucleophile in families GH33 and GH34, nor possess a carboxylate-containing residue that could also fulfil this role. Very recently it has been shown for the GH83 enzymes at least, that these endo-sialidases hydrolyze with inversion of configuration.47 As there is only one suitably positioned carboxylate-containing residue in the active site, it has been proposed this residue acts as the general acid, and the carboxylate moiety in the sialic acid substrate acts as the general base; although this mechanism has yet to be verified in a number of enzymes, it does suggest another, completely independent, substrate-assisted mechanism is employed by glycosidases. It has also been shown in recent years that glycosidases in families GH4 and GH109, which both hydrolyse with retention of configuration, use an unusual NAD+-dependent mechanism and that the GH4 enzymes also require a divalent metal ion.24,25 NAD+ is believed to abstract a hydride from the C3 carbon atom of the glycoside to form a ketone, which acidifies the proton at the C2 position and allows deprotonation by a residue acting as a base. This is accompanied by elimination of the glycosidic oxygen by a residue acting as an acid, leading to a 1,2-unsaturated intermediate that is stabilised by the metal ion (in GH4 enzymes). A water molecule attacks at the anomeric position, following base activation, to generate a 3-keto-glycoside, which is subsequently reduced by NADH to regenerate NAD+.
What makes a good mechanism-based glycosidase inhibitor?
A great deal of attention over the past decade has been paid to increasing the potency of glycosidase inhibitors with a view to designing drugs which could be used in a plethora of medical applications. In this review, we will not be discussing any inhibitors which bind covalently to the enzyme, but this has been extensively reviewed recently by Rempel and Withers.48 In many cases inhibitors are proposed to mimic the transition state formed fleetingly during glycoside hydrolysis, but this has been demonstrated conclusively in very few cases. Wolfenden has shown glycoside-degrading enzymes are able to enhance the rate of catalysis 1017 fold over the uncatalysed reaction, making them a remarkably proficient class of enzyme.3 This would translate to an estimated dissociation constant for the transition state to not be greater than 10−22 M, indicating that the transition state binds to an enzyme with extraordinary affinity.3,49 Pauling first postulated in the 1940s that the highest affinity inhibitors for an enzyme were likely to be those that mimicked the structure of a ‘strained activated complex’![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50,51 (i.e. transition state). In the case of glycoside hydrolysis, the short-lived transition state possesses substantial oxocarbenium character (Fig. 2a and b); it consists of partially formed or broken bonds between the attacking nucleophile (which may be a deprotonated water molecule, carboxylate group, acetamido group or a tyrosine residue) and the glycosidic oxygen of the leaving group (or the attacking water molecule in the deglycosylation step of a retaining mechanism). Under these conditions the anomeric carbon possesses trigonal character, which causes sp2 hybridisation predominantly along the bond between the anomeric carbon and endocyclic oxygen.34 The double bond character imposed on the pyranose ring means there is distortion from a relaxed chair conformation typical of the substrate, which causes C1, C2, O5 and C5 to lie in a plane and to take a more energetically unfavourable half chair or boat conformation at, or close to, the transition state (Fig. 2c).34,52 There is also a considerable build-up of positive charge on the pyranose ring at the transition state, which is delocalised along the bond between the anomeric carbon and endocyclic oxygen.
50,51 (i.e. transition state). In the case of glycoside hydrolysis, the short-lived transition state possesses substantial oxocarbenium character (Fig. 2a and b); it consists of partially formed or broken bonds between the attacking nucleophile (which may be a deprotonated water molecule, carboxylate group, acetamido group or a tyrosine residue) and the glycosidic oxygen of the leaving group (or the attacking water molecule in the deglycosylation step of a retaining mechanism). Under these conditions the anomeric carbon possesses trigonal character, which causes sp2 hybridisation predominantly along the bond between the anomeric carbon and endocyclic oxygen.34 The double bond character imposed on the pyranose ring means there is distortion from a relaxed chair conformation typical of the substrate, which causes C1, C2, O5 and C5 to lie in a plane and to take a more energetically unfavourable half chair or boat conformation at, or close to, the transition state (Fig. 2c).34,52 There is also a considerable build-up of positive charge on the pyranose ring at the transition state, which is delocalised along the bond between the anomeric carbon and endocyclic oxygen.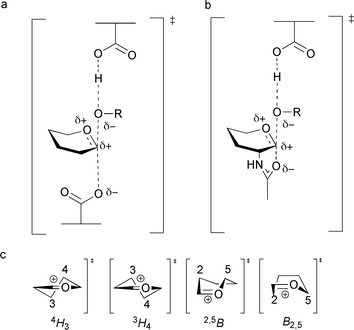 | ||
| Fig. 2 Structure of the oxocarbenium ion-like transition state, formed during glycoside hydrolysis of (a) the glycosylation step of the ‘classical’ retaining mechanism and (b) the glycosylation step of substrate-assisted catalysis; R is the leaving group. (c) Possible transition state conformations employed during glycoside hydrolysis (half chair (4H3 or 3H4) or boat (2,5B or B2,5) conformations). | ||
There are numerous examples of glycosidase inhibitors that have been designed, synthesised or tested as transition state mimics. This is largely based on the incorporation of one or more features, such as charge or geometry, which mirrors that proposed for the transition state in question. Some would argue that the classification of these inhibitors as “transition state analogues” is not wrong, as, at an intuitive level the features incorporated in an inhibitor may mimic the transition state. However, to further improve potency of transition state analogues, with the aim of harnessing more of the transition state binding potential, it should be ascertained which are actually true analogues in an enzyme system as opposed to those which bind fortuitously. This is obviously difficult to define, but a number of criteria have been proposed and some are discussed below.
One approach for studying transition state mimicry involves investigating linear (Gibbs) free energy relationships (LFERs). For a true transition state mimic, alterations in chemical structure (of the substrate and inhibitor) or the surrounding environment (of the enzyme) should have an equal effect on enzyme rate enhancement and the affinity of the inhibitor relative to the substrate. Thus, for a true transition state analogue, the inhibition constant (Ki) is proportional (by a ‘proportionality constant’x) to the dissociation constant for the transition state. Wolfenden and Thompson (independently) first alluded to this relationship for classifying peptide aldehyde inhibitors as transition state analogues of papain and elastase, respectively.53,54 If alterations are made to the enzyme (by site-directed mutagenesis to active site residues that are not vital to catalysis) or equivalent structural changes are made to a series of substrates and inhibitors so that the uncatalysed reaction rate remains unchanged, then a plot of log KM/kcat against log Ki should yield a straight line with a slope equal to 1 if the inhibitor mimics the transition state.55 The correlation does not indicate the degree to which an inhibitor mimics the transition state, but a slope of 1 signifies that an inhibitor (at least the part that has been altered or tested by a mutation in the enzyme) is a transition state analogue (this is well described in Ref. 55). The relationship also assumes there is no change in the rate determining step of the enzyme-catalysed reaction.55
Wolfenden and co-workers have demonstrated that enzymes lower the enthalpy of activation for a reaction substantially, and the degree to which the enthalpy of activation is lowered correlates with the effectiveness of the enzyme as a catalyst. The reliance of enzymes on a substantial enthalpy of activation for the most proficient catalysis means that rate enhancements increase considerably with decreasing temperature.56,57 It follows that the increased affinity for the transition state over the ground state substrate by an enzyme is therefore largely enthalpic in origin. Wolfenden has therefore proposed that it is reasonable to assume that inhibitors which truly mimic the transition state would also derive their binding from large enthalpies of activation. Also, as rate enhancements increase considerably as the temperature is decreased for these proficient enzymes, the enzyme affinity for inhibitors that mimic the transition state should also increase more sharply with a decrease in temperature than those that are ground state substrate analogues.56,57
Some elegant examples of how the predicted structure of the transition state can influence inhibitor design are illustrated by the work of Schramm and colleagues using a combination of kinetic isotope effects, computer modelling, X-ray crystallography and NMR spectroscopy.58 The work stresses the need for mimicry of the transition state shape and volume, examination of the molecular electrostatic potential surface similarity between the transition state and putative analogues, and consideration of the energy that every functional group in the transition state contributes to binding with the enzyme. These methods have been employed in the design of extremely potent inhibitors for human purine nucleoside phosphorylase,59,60 the inhibition of which has great therapeutic potential; indeed, these compounds are currently in clinical trials for the treatment of T-cell cancers and autoimmune disorders.61 Similar techniques were used in the inhibitor design of a 5′-methylthioadenosine/S-adenosylhomocysteine nucleosidase, which yielded inhibitors in the low femtomolar range.62,63 It is interesting to note that despite significant homology between enzymes from different species, the transition state “poise” varied subtly which aided the design for both potency and specificity, and has ramifications for inhibitor design that may be isozyme, tissue or species specific.58
Glycosidase inhibitors
The number of putative glycosidase inhibitors is continually growing, and it is outside the scope of this review to mention them all in detail. Instead, we will describe the general features of representative glycosidase inhibitors from different classes (concentrating on those that possess six-membered rings and largely omitting the body of work that has been done on the pyrrolidines and pyrrolizidines), which will be used to put the later discussion about the properties of transition state mimics in context; more comprehensive and detailed reviews from the last decade about the inhibitors themselves can be found in Ref. 64–70. By far the most popular way of designing glycosidase inhibitors is to incorporate a nitrogen atom in the saccharide ring and in many cases Nature beat man to the synthesis. Nojirimycin 1 (Fig. 3) was the first natural saccharide mimic to be isolated from a Streptomyces strain in 1966,71 and was originally proposed to contain antibiotic properties. Although capable of inhibiting both α- and β-glycosidases, the hydroxyl group at C1 of the imino sugar is unstable.72 The synthesis of deoxynojirimycin 2, which lacks this hydroxyl group, was accomplished the following year by Paulsen and colleagues,73 and also later shown to occur naturally.74,75 Deoxynojirimycin served as a template for the subsequent synthesis of different glycoside derivatives (some of which also occur in Nature), including deoxymannojirimycin and deoxygalactonojirimycin, and all tend to be more potent inhibitors of α-glycosidases than β-glycosidases.76 1,2-Dideoxynojirimycin, or fagomine, was also isolated,77 but was a less potent inhibitor.78 The drug Miglitol 3,15 used in the treatment of type II diabetes (described earlier), builds on the deoxynojirimycin scaffold with a hydroxyethyl group at the nitrogen atom and Zavesca/Miglustat 4,79,80 used to combat the symptoms of Gaucher disease (by inhibition of the glycosyltransferase biosynthesizing the glucosylceramide substrate which accumulates during the disease), possesses an N-butyl group.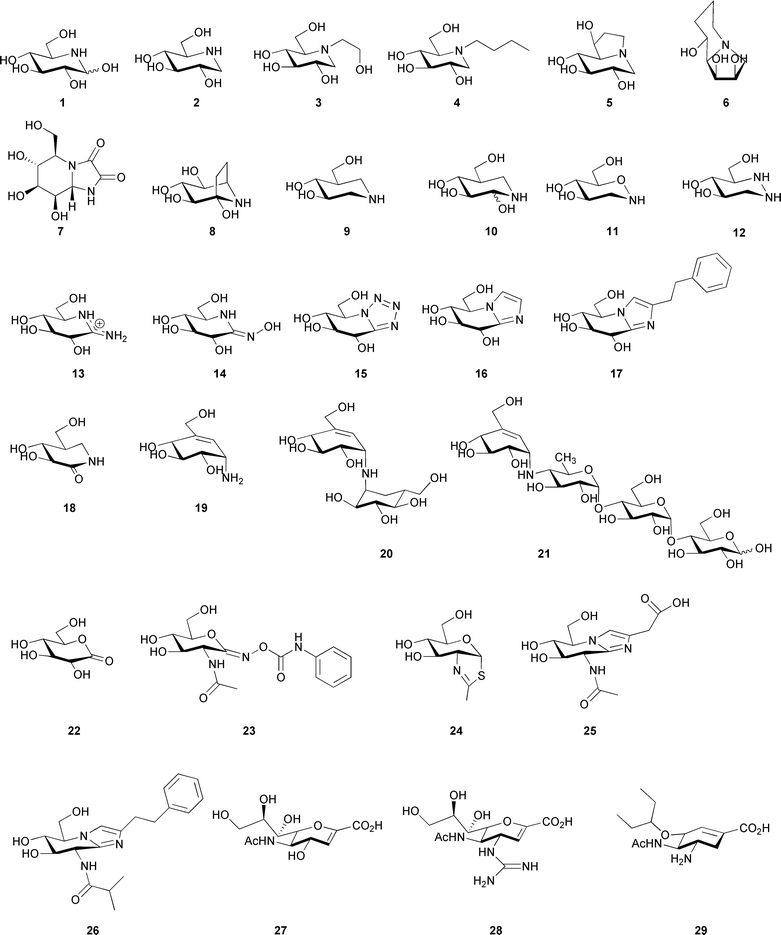 | ||
| Fig. 3 Structure of nojirimycin (1), deoxynojirimycin (2), Miglitol (3), N-butyl deoxynojirimycin (4), castanospermine (5), swainsonine (6), kifunensine (7), calystegine B2 (8), isofagomine (9), noeuromycin (10), tetrahydrooxazine (11), azafagomine (12), gluco-amidine (13), gluco-hydroximolactam (14), glucotetrazole (15), unsubstituted glucoimidazole (16), phenethyl-substituted glucoimidazole (17), isofagomine lactam (18), valienamine (19), validoxylamine A (20), acarbose (21), gluconolactone (22), PUGNAc (23), NAG-thiazoline (24), gluco-nagstatin (25), GlcNAcstatin C (26), 2-deoxy-2,3-didehydro-N-acetylneuraminic acid (27), Relenza (28), and Tamiflu (29). | ||
Many ideas for the synthesis of indolizidines, where a five and a six membered ring are fused together, also came following the discovery of natural products. Castanospermine 5, first isolated from the legume Castanospermum australe81 and later chemically synthesised,82 can inhibit both α-glucosidases and β-glucosidases, and may play a role in preventing spread of dengue virus infection by inhibiting an endoplasmic reticulum α-glucosidase.83 Similarly swainsonine 6, isolated from Swainsona canescens84 and later synthesised,85 and kifunensine 7, which was isolated from Kitasatosporia kifunense86 and also chemically synthesised,87 are both potent α-mannosidase inhibitors;88,89 the latter has shown promise as an anticancer agent.90 Calystegine compounds, which possess a nortropane ring system were first discovered in the early 1990s as natural products in the roots of Calystegia sepium91 and subsequently a number have been chemically synthesised (reviewed in Ref. 92). They are a complex family of molecules with a naming system based on the position of the nitrogen atom and the configuration of the hydroxyl groups, and many inhibit glycosidases with different specificities; calystegine B28 is shown here as a representative example. Although 8 is pseudo-symmetrical with respect to its hydroxyl groups, when visualised in complex with a β-glucosidase (Fig. 4a) it was observed in an orientation similar to isofagomine 9 (see below) rather than deoxynojirimycin 2.93,94
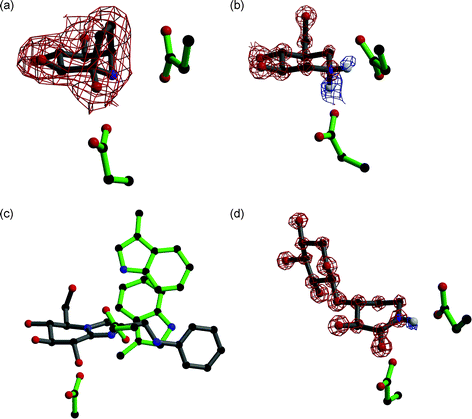 | ||
| Fig. 4 (a) Calystegine (8) in complex with a family 1 β-glucosidase (PDB code 2CBV94); the residue below the inhibitor is the catalytic nucleophile and the residue to the right is the acid/base. Observed electron density (for the maximum likelihood weighted 2Fobs–Fcalc map, contoured at 1σ) is shown for calystegine, showing it binds in a similar orientation to isofagomine. (b) Cellobio-derived form of isofagomine (9) in complex with a family 5 endoglucanase (PDB code 1OCQ101); the residue below the inhibitor is the catalytic nucleophile and the residue to the right is the acid/base. Observed electron density for the maximum likelihood weighted 2Fobs–Fcalc map, contoured at 2.5σ, is shown in red and for the Fobs–Fcalc map, contoured at 2.1σ, is shown in blue. The ‘difference’ density shows the presence of two hydrogen atoms on the nitrogen atom of isofagomine. (c) Phenylaminomethyl-substituted glucoimidazole in complex with a family 3 β-D-glucan glucohydrolase (PDB code 1X39133); the residue below the inhibitor is the catalytic nucleophile and the residue to the right is the acid/base. The two tryptophan residues in the active site are proposed to make hydrophobic interactions with the phenyl ring of the inhibitor, but this interaction has not been observed in all enzyme complexes with substituted imidazole inhibitors. (d) Xylobio-derived isofagomine lactam in complex with a family 10 xylanase (PDB code 1OD8137); the residue below the inhibitor is the catalytic nucleophile and the residue to the right is the acid/base. Observed electron density for the maximum likelihood weighted 2Fobs–Fcalc map, contoured at 4σ, is shown in red and for the Fobs–Fcalc map, contoured at 1.8σ, is shown in blue. The ‘difference’ density shows the presence of a hydrogen atom on the nitrogen atom of the isofagomine lactam, indicating it exists as the amide tautomer and not the iminol as originally proposed. | ||
Pioneering work in the Bols group in the early 1990s,95–97 and later also by Ichikawa and co-workers,98–100 saw the synthesis of isofagomine 9 and its derivatives. Isofagomine was the first compound to possess a nitrogen atom in place of the anomeric carbon. It tends to inhibit β-glycosidases more potently than α-glycosidases, which may be a consequence of charge accumulation at different positions in the transition state for the different enzymes. Interestingly, the atomic resolution structure of the cellobio-derived form of 9 in complex with an endoglucanase showed the secondary amine functional group was protonated and both catalytic glutamate residues were deionised101 (Fig. 4b). Although 9 is a potent glycosidase inhibitor, it was predicted that the inclusion of a hydroxyl group at the C2 position would increase this potency as this group had been shown to contribute considerable binding energy to stabilisation of the transition state, predominantly through interactions with the nucleophile.102 Although synthesised as a mixture of anomers at C2, noeuromycin 10 strongly inhibits both α- and β-glycosidases.97,103 Another isofagomine analogue, tetrahydrooxazine 11, which was also synthesised by Bols and co-workers,104 possesses an endocyclic oxygen atom as found in native glycosides which enables additional hydrogen bonding potential that is unavailable in 9 and 10. Although a reasonable inhibitor of a β-glucosidase, its lower potency than 9 may be a consequence of the weak basicity of the nitrogen atom which is unlikely to be protonated. An attempt to overcome this came in the form of azafagomine 12.105–107 Azafagomine 12 incorporates two nitrogen atoms, and thus combines features of both 2 and 9; it is possible that either nitrogen atom may be protonated and as such may better reflect the charge distribution at the transition state.
The inhibitors discussed so far are typically found in a relaxed chair conformation in solution, although some distortion may occur upon binding to an enzyme. The majority of them possess a protonated secondary amine group (although this has only been shown unequivocally for 9101), and hence to some degree mimic the charge distribution at the transition state. A different approach is to incorporate group(s) in the inhibitor which causes distortion of the compound to mimic the geometry of the transition state; this may also introduce a specificity element as different enzymes use itineraries via transition states with varying conformations.52 Amidine inhibitors, such as the gluco form 13, were synthesised in the Ganem group in the early 1990s,108 followed by amidrazone and amidoxime (also called hydroximo-1,5-lactam or 1,5-lactam oxime) derivatives, in gluco14, manno and galacto forms,109,110 which were shown to inhibit both α- and β-glycosidases.110,111 A number of functional groups have been added to the exocyclic nitrogen in order to mimic the aglycon, both with the amidine112-114 and amidoxime/hydroximolactam115,116 inhibitors. In addition the latter has also been synthesised in an oligosaccharide form.117 The conjugation of the carbon–nitrogen double bond of the hydroximolactam group (which has been shown to be exocyclic) and the endocyclic nitrogen atom causes distortion of the ring to a half chair conformation which may mimic the geometry at the transition state.115 Consistent with protonation by the acid/base residue using a lateral protonation mechanism, these inhibitors were synthesised on the premise of providing a heteroatom in the ring plane which should allow a strong hydrogen bond interaction with the acid/base residue.67
Vasella and colleagues pioneered the synthesis of glucotetrazole 15 in the early 1990s, to create a new class of inhibitors related to 2.118 Subsequently other derivatives were developed,119,120 which all proved to be good β-glycosidase inhibitors. The addition of the tetrazole moiety fused onto the pseudo-glycoside ring causes distortion of the ring into a half chair conformation.118 Along similar lines, imidazole (or tetrahydroimidazopyridine) derived compounds, the idea of which came from the naturally occurring inhibitor nagstatin (see below), was again first developed in the Vasella group; the unsubstituted gluco form 16 is shown in Fig. 3. These compounds have been synthesised to mimic different glycosides,121-124 with a number of different functional groups attached to mimic the aglycon,125-129 and in oligosaccharide forms.117 It is interesting to note that if a triazole moiety is fused to the pseudo-glycoside ring (i.e. the nitrogen atom adjacent to the anomeric carbon is replaced with a carbon atom), then extremely weak inhibitors result. This supports the notion that the heteroatom attached to the anomeric carbon makes an important interaction with the acid/base residue; the lone pair of electrons for this interaction is in the plane of the ring suggesting that protonation of the glycosidic oxygen also occurs in this manner.130 The imidazole compounds are more basic than the tetrazole and triazole compounds, however, which has been shown to correlate with greater inhibition.124 Compound 16 is a highly potent inhibitor of β-glucosidases, but a weaker inhibitor of α-glucosidases.121 Atomic resolution analysis with the cellobio-derived form of 16 indicated a strong hydrogen bond interaction between the nitrogen atom adjacent to the anomeric carbon and the acid/base residue; the protonation trajectory was in the plane of the imidazole ring, and the proton was observed to lie closer to the nitrogen atom of the imidazole. Glucoimidazole compounds with the addition of functional groups were synthesised on the premise of increasing potency mimicking the aglycon and thus taking advantage of interactions in the +1 subsite125 (for subsite nomenclature see Ref. 131). These groups did indeed improve potency in many cases, with Ki values as low as 0.11 nM for the phenethyl-substituted glucoimidazole 17.125 The general mode by which these imidazole derivatives gain potency, however, remains contentious; although the observations of phenyl- and phenylaminomethyl-substituted glucoimidazole in complex with a family 3 β-D-glucan glucohydrolase (Fig. 4c), and the manno form of 17 in complex with a family 2 β-mannosidase, indicated there were interactions between the phenyl group and tryptophan residues in the active site of the enzyme,132–134 a similar study with two family 1 β-glucosidases in complex with 17 showed an absence of any such interaction.135
Distortion of the ring to generate compounds which may resemble the geometry of the transition state can also be done by introducing a double bond in the pseudo-glycoside ring itself. Isofagomine lactam, the gluco form 18 is shown in Fig. 3, the synthesis of which was first reported by Williams et al., was proposed to exist as the iminol tautomer where a double bond would exist in the pseudo-glycoside ring with a hydroxyl group at C2.136 Atomic resolution crystallography later disproved this theory and showed it existed as the energetically favoured amide tautomer137 (Fig. 4d). A number of different glycoside forms of isofagomine lactam have since been synthesised, and have proved to be potent inhibitors of β-glycosidases.138 Valienamine 19, and its analogues, have also proven to be potent inhibitors primarily of α-glycosidases.139 Valienamine was first discovered as a product of validoxylamine A hydrolysis by Pseudomonas denitrificans, which in turn was a product of validamycin A hydrolysis,140 and was later chemically synthesised.141 Validoxylamine A14220 is a pseudo-disaccharide which proved to be a potent trehalase inhibitor;143 the first structure of a trehalase was solved recently and demonstrated the valienamine unit bound in the −1 subsite, and the pseudo-glucose in the +1 subsite.144 The valienamine unit is also found in acarbose 21,14 a drug which targets α-glucosidases in the treatment of type II diabetes (described earlier). The valienamine part of the compound is usually found in the −1 subsite when visualised in complex with an enzyme, such as a glucoamylase,145 suggesting it may act as a transition state mimic.
One of the most appealing ways to design a transition state analogue would be to incorporate both the features of geometry and charge present at the transition state. Glyconolactones, such as gluconolactone 22, which unlike many inhibitors possess an endocyclic oxygen atom as in a native glycoside, were first shown to inhibit β-glycosidases (but to a lesser extent α-glycosidases) in the early 1940s.146,147 The conjugation in the glyconolactones was predicted to cause distortion to a conformation typical of the transition state;148 the gluco- and galactonolactone were later observed in a half chair conformation when in complex with an enyzme.149,150 It has also been postulated that the charge distribution of the compounds may mimic the partial positive charge at the transition state.151 Unfortunately, however, these compounds are unstable as they are prone to ring opening, which makes their study as glycosidase inhibitors limited.152 Attempts have also been made to include sulfur in the pseudo-glycoside ring of a number of different glycoside forms, as well as the sulfoxide or sulfone derivatives. In the majority of cases, however, these have proved to be weak inhibitors of both α- and β-glycosidases.153,154 Inhibitors containing a sulfonium ion, where the positively charged sulfur atom in the pseudo-glycoside ring may mimic the charge at the transition state, have been successfully synthesized. Elucidation of the structure155 and subsequent synthesis156,157 of the naturally occurring salacinol demonstrated these compounds were viable, and their inhibition of α-amylases158 suggested they may be used in the treatment of diabetes. Subsequently a number of analogues of salacinol have been synthesized159 and other known glycosidase inhibitors have been modified to possess a sulfonium ion.160,161
Enzymes which catalyze substrates containing N-acetyl hexosamine using a substrate-assisted mechanism are amenable to the design of selective and potent inhibitors which incorporate features designed to mimic the transition state which exists on the way to and from an oxazoline ion intermediate. Early work in the field focussed on PUGNAc 23,162 which possesses an N-phenylcarbamate moiety that makes a number of interactions with active site residues.21,163 The deduction of the mechanism led to the design of NAG-thiazoline 2439 and the subsequent quest for selectivity provided a number of other derivatives with different lengths of chains19 and groups37 at the N-acyl position, many of which proved to be extremely potent inhibitors. A number of other inhibitors incorporate some of the features already described, combined with the GlcNAc functionality; for example gluco-configured nagstatin 25,164 which is based on the naturally occurring nagstatin and provided the basis for the synthesis of the imidazole derivatives. This was developed further into a PUGNAc-imidazole hybrid164 by the Vasella group and a series of GlcNAcstatins, such as GlcNAcstatin C 26165,166 by van Aalten et al.; the incorporation of an imidazole moiety in these compounds causes distortion of the ring to a geometry which may resemble the transition state.
Inhibitors targeting sialidases have received a great deal of interest over the past decade or so because of the vital role the enzyme plays in the life cycle of the influenza virus and thus the spread of the disease.167 Early work in the field focussed on the naturally occurring 2-deoxy-2,3-didehydro-N-acetylneuraminic acid 27,168 which contains a double bond in the pseudo-glycoside ring that may mimic the geometry at the transition state. Structure-based drug design methods were used to replace the hydroxyl group at the C4 of 27 with a guanidino group, which lead to the sub-nanomolar inhibitor Relenza 28.16 Further developments were attempted in order to make a drug with better oral bioavailability than Relenza, which was achieved by changing the glycerol moiety to a group with more hydrophobic character.18 This led to the drug now known as Tamiflu 29 which is presently used world-wide in the treatment of the influenza virus.
Many glycosidases hydrolyse substrates longer than those which involve a single monosaccharide being present in the −1 subsite, and indeed for a large number it is an absolute requirement. This adds complexity to the study of glycosidase inhibitors, as for many enzymes an inhibitor which is similar in size to a monosaccharide does not provide enough binding potential for the compound to be analysed as an inhibitor; additional residues are required in the other glycon and/or aglycon binding sites for this potential to be realised. If the synthesis of these single unit inhibitors was not enough of a challenge for chemists, the addition of further sugars onto them is approaching a near impossibility. Nevertheless, in a number of cases this has been achieved. Glycosynthases169 have been used to generate cellobio-, cellotrio- and cellotetrao- versions of isofagomine 9 and tetrahydrooxazine 11; in each case the inhibitors became more potent as the length of the inhibitors increased demonstrating the requirement for the binding in the −2, −3 and −4 subsites.170 Others have used a synthetic approach to obtain cellobio- and cellotrio- analogues containing a hydroximolactam (14), tetrazole (15) or imidazole (16) unit. These studies demonstrated that some inhibitors benefited from having additional sugars at the non-reducing end, while others had an improved potency when on the reducing end (where the sugars would lie in the product subsites).117 Xylobio- derivatives of 2, 9, 14 and 16 have also been chemically synthesised.171 In recent years the synthesis of oligosaccharides where the glycosidic oxygen is replaced by a sulfur atom has also become extremely useful at providing inhibitors which cannot be hydrolysed by the enzyme.172
Transition state mimicry or fortuitous binding
Given the volume of publications on glycosidase inhibitors, only a handful have addressed the issue of whether they are mimics of the transition state or fortuitous binders. These studies have been extremely insightful, and although they involve a large volume of experiments and analysis, should be encouraged for other enzyme systems and inhibitors in the future. We will now describe the conclusions from these studies, classified by a number of criteria proposed for true transition state mimics.Linear (Gibbs) free energy relationships (LFERs)
As alluded to earlier, one approach for studying whether an inhibitor is a true transition state mimic is by measuring LFERs. Thus, for a true transition state mimic, changes in chemical structure of the substrate and inhibitor or the surrounding environment of the enzyme should have an equal effect on enzyme rate enhancement and the affinity of the inhibitor relative to the substrate.55 In real experimental terms, this means that corresponding changes in series of substrates and inhibitors or a series of mutational alterations in the enzyme (that are not vital for catalysis) should influence the KM/kcat and Ki to the same extent. Note that this does not demand that the inhibitor possesses every interaction present at the transition state, for it is the relative changes in inhibitor and transition state binding that are assessed. The LFER technique has been applied successfully to a number of studies examining the transition state analogy of peptidase inhibitors.173–176To our knowledge the first time a full LFER study was conducted for a glycosidase was by Ermert et al. who examined the gluco- and manno-derived forms of tetrazole119 (for the gluco-form see 15), although Field and colleagues had considered correlations between Ki and KM/kcat, albeit in a more crude form, in 1991.177 Ermert et al. measured the catalytic rates on two synthetic substrates using a range of enzymes (α-mannosidases, a β-mannosidase, an α-glucosidase, a β-glucosidase and a β-galactosidase) and the inhibition of both tetrazole compounds on these same enzymes. The methods used here are not totally in accordance with the way in which LFERs are usually conducted as the authors chose to test the compounds in a number of different enzymes. It has, however, been shown that different enzymes use various conformation itineraries52 and even very closely related enzymes may have different transition state poise,60,178 and so what may be a good transition state mimic for one class of enzymes may not be so for others. Although it is not entirely clear in the publication which of the inhibition data reported are included in the correlation of log 1/Ki against log kcat/KM, the plot nevertheless gives a linear correlation with a slope of 1.1,119,179 which suggests tetrazole-derived inhibitors are good transition state mimics. The distortion caused by the fusion of the tetrazole moiety to the pseudo-glycoside ring into a half chair (or envelope) conformation may therefore closely represent the structure at the transition state.
A LFER study of the tetrasaccharide inhibitor acarbose (21) was performed by Mosi et al. using a number of active site mutants of the GH13 cyclodextrin glycosyltransferase.180 The study was conducted using two different synthetic substrates for the enzyme (which differed in length and hence number of subsites occupied), and the dependence of log Ki against both log KM/kcat and log KM was assessed. In each case the plot yielded a reasonable correlation; the log Kivs. log KM/kcat plot gave slopes of 2.2 and 1.6 for the ‘monosaccharide’ and ‘trisaccharide’ substrates, respectively, whereas the plot for log Ki against log KM gave slopes of between 0.4 and 0.5. The correlation was better (r2 of 0.96 vs. 0.81) for the ‘monosaccharide’ substrate, which was assumed to be a better test for the −1 subsite where the valienamine unit of acarbose lies; this comparison does, however, show the influence of the choice of substrate upon LFER studies in general. The authors interpreted the data as acarbose displaying ‘equal mimicry of the ground state and transition state’.180 This, however, is at odds with the derivation of the equation underpinning the use of LFERs, where a transition state analogue is defined only when there is a direct correlation between log Ki and log KM/kcat with a slope of 1.55 An independent study by Berland et al. used kinetic methods to determine the kcat/KM parameters for a series of active site mutants of a GH15 glucoamylase and isothermal titration calorimetry (ITC) methods to measure the Kd values for acarbose.181 The plot of log Kd and log KM/kcat once again gave a reasonable correlation, with a slope of 2. The authors accounted for the fact the correlation should have a unity slope, and concluded that acarbose is not a true transition state mimic for this enzyme.181 The double bond in the valienamine unit of acarbose may prevent the appropriate ring distortion to mimic the transition state, but nevertheless remains a highly potent inhibitor. During the same study deoxynojirimycin (2) was also studied as a candidate for a transition state analogue, but no correlation was seen between the kinetic and inhibition parameters which suggests it is not a true mimic of the transition state.181
Whitworth et al. examined LFERs with two inhibitors which target enzymes that use a substrate-assisted mechanism.182 They used an elegant system of modifying the volume of the alkyl group of the N-acyl moiety at the C2 position in both the substrates used to monitor the reaction and the inhibitors tested: PUGNAc (23) and NAG-thiazoline (24). The experiments were conducted on the human O-GlcNAcase, which is classified into family GH84. The resulting plots of log 1/Ki against log kcat/KM for the NAG-thiazoline series showed a strong correlation with a slope close to 1 suggesting it is a true transition state mimic. The opposite, however, was true for the PUGNAc series, which show a weaker correlation suggesting this compound is less likely to mimic the transition state.182 The results were perhaps surprising given the sp2 anomeric centre in PUGNAc, which may have been predicted to resemble the transition state more than the equivalent sp3 centre in NAG-thiazoline, which may have been expected to better represent the oxazoline intermediate (see Fig. 1). The authors postulated that these unexpected results may be attributable to the longer C–S bond length present in NAG-thiazoline in comparison to the C–O bond in the oxazoline intermediate.182 The longer bond length may mimic the partial, but significant, bond order between the nucleophilic carbonyl oxygen atom and anomeric carbon of a late transition state.
It is intriguing that the general conclusions in terms of sp2- and sp3-centred inhibitors drawn from a LFER study by Wicki et al. were almost opposite to those described for PUGNAc and NAG-thiazoline (although they were tested against different enzymes which may have significantly different transition states). Wicki et al. used a series of active site mutants (both ‘first sphere’ and ‘second sphere’) of a xylanase to assess the transition state analogy for the xylobio-derived versions of deoxynojirimycin (2), isofagomine (9), lactam oxime (14), imidazole (16) and isofagomine lactam (18).183 The log kcat/KM against log 1/Ki plot for each inhibitor showed a slope of close to 1, but the correlation coefficients for the sp3-centred inhibitors (those which were more likely to resemble the charge at the transition state rather than the geometry) deoxynojirimycin and isofagomine were poor (r values of 0.89 and 0.77, respectively, which equates to r2 values of 0.79 and 0.59 to allow direct comparison with values described below). However, the sp2-centred inhibitors (which were more likely to resemble the geometry at the transition state) all showed a correlation coefficient very close to 1, suggesting they were transition state mimics.183 These data are in agreement with the earlier study by Ermert et al. on the tetrazole inhibitors, and suggest that those inhibitors which can distort the pseudo-glycoside ring into a half chair or envelope conformation may generally be the best mimics of the transition state.
A detailed investigation into the LFERs of 8 mannosidase inhibitors was performed by Tailford et al. with a series of active site mutants of a family 2 mannosidase.134 In particular this study examined a number of manno-derived imidazoles (see 16 for gluco unsubstituted version) and amidines (see 13 for gluco unsubstituted version) with a number of functional groups attached which may mimic the aglycon; all compounds were observed in a B2,5 conformation in the active site of the enzyme. The substituted manno-imidazole compounds (the three tested had phenethyl (see 17 for the gluco version), phenylaminomethyl and phenyloxymethyl functional groups) all showed a strong correlation in the plot of log KM/kcat against log Ki with a slope close to 1 (Fig. 5a) suggesting they were transition state analogues. The plot for the unsubstituted manno-imidazole also had a slope close to 1, but the r2 correlation was lower (at 0.78).134 The study by Wicki et al. showed the LFER plot for the equivalent xylobio-derived compound had a high correlation coefficient suggesting it was a good transition state analogue;183 this may, however, reflect the differences in the structure of the transition state for different enzyme families. This observation also suggests that, for this enzyme, the presence of the functional group in the aglycon was required to properly mimic the transition state. The two substituted amidines tested, isoquinuclidine and noeuromycin 10, all showed slopes of less than one and poor correlation coefficients, suggesting none of them were good mimics of the transition state.
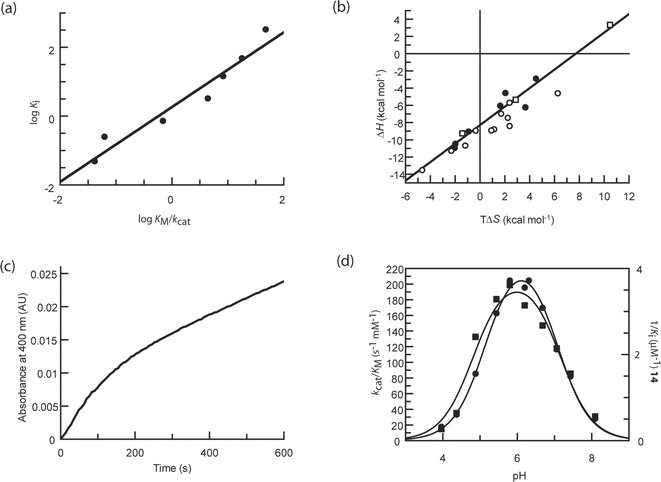 | ||
| Fig. 5 (a) Linear free energy plot of log Ki against log KM/kcat for the phenethyl-substituted mannoimidazole with the family 2 β-mannosidase (data taken from Ref. 134). The best fit line through the points has a slope of 1.09 and a correlation r2 of 0.94, which strongly suggests the compound is a good mimic of the transition state. (b) Enthalpy–entropy compensation plot for inhibitors with a β-glucosidase; the line of best fit has a slope of 0.93 and correlation of 0.91. Points in circles represent data from the initial study of 18 compounds (data taken from Ref. 184); filled circles represent those which mimic the charge at the transition state, open circles those which mimic the geometry. The points in squares represent data from subsequent studies (data taken from Ref. 194 and 195) and demonstrate that even when a compound is enthalpically unfavourable, it still falls on the line. (c) Plot of gain of product (as indicated by an increase in absorbance at 400 nm by production of 2,4-dinitrophenolate from hydrolysis of 2,4-dinitrophenyl β-D-glucopyranoside by a β-glucosidase) against time to illustrate slow onset inhibition. Slow onset inhibition is characterised by an initial rapid catalytic rate (here for 200–300 s), followed by a slower steady state rate. (d) pH dependence of kcat/KM for a β-glucosidase (circles) and 1/Ki for 14 (squares); fits to bell-shaped ionisation profiles are shown (data taken from Ref. 184). In this case the pH dependence for inhibition mirrors that of catalysis, but this is not the case for all inhibitors. | ||
In contrast to definitions of transition state mimicry which depend solely on the intuition or prejudice of an individual scientist, LFERs produce a quantifiable measurement that others can repeat. This does not mean, however, that LFER analyses are without flaws. Firstly, if the goal of the study is to simply obtain tight-binding enzyme inhibition, then a LFER correlation is irrelevant; it would be a mistake to be discouraged by a poor LFER performance if one is primarily only interested in Ki values. The second issue, especially where site-directed mutagenesis is the perturbation of choice in the LFER study, is the selection of the mutations made. Some would argue, for example, that charged inhibitors do comparatively poorly in LFER analysis (discussed in Ref. 134) because the most energetically significant interactions of these charged compounds are with the nucleophile and acid/base residues; the two residues which cannot be mutated in the analysis without compromising the enzyme activity completely. One can imagine that, were it possible, an LFER plot which included nucleophile and acid/base variants would show massive disruption to catalysis and a similar reduction in inhibitor binding. This raises the issue of how to address the compounds that do “poorly” in LFER tests. Do they mimic something other than the transition state, are they fortuitous or adventitious binders, or has the technique itself legislated against certain classes of compounds?
Thermodynamics of inhibitor binding
As described earlier, Wolfenden proposed that the increased affinity for the transition state over the ground state by an enzyme is primarily derived from enthalpy, and therefore it could be argued that inhibitors which truly mimic the transition state should also bind with large enthalpic contributions.56,57 The issue of which glycosidase inhibitors were likely to be transition state mimics based on their binding thermodynamics was tackled using a panel of 18 inhibitors with a family 1 β-glucosidase.184 Measurements were made using ITC, which gives a direct read-out on the affinity of the inhibitor, stoichiometry of binding between inhibitor and enzyme and the enthalpic contribution to binding, from which the Gibbs free energy and entropy can be calculated (for a recent review on ITC see Ref. 185). All of the inhibitors examined in this study (deoxynojirimycin 2, castanospermine 5, calystegine B28, isofagomine 9, noeuromycin 10, tetrahydrooxazine 11, azafagomine 12, gluco- 14 and galacto-hydroximolactam, glucotetrazole 15, unsubstituted glucoimidazole 16, glucoimidazole substituted with a phenethyl group 17 and a range of other functional groups, and isofagomine lactam 18) displayed a negative and therefore favourable enthalpic contribution to binding.184 Some of the inhibitors, however, harnessed a significant portion of their binding energy from entropy, which questions their ability to be transition state mimics. These include the phenethyl-substituted glucoimidazole 17 (which incidentally was the most potent of the inhibitors examined in the study), isofagomine 2, and calystegine B28. The data show there is no correlation between the inhibitor affinity and the enthalpic contribution to binding, and nor does the chemistry of the inhibitor appear to influence the thermodynamics, at least not on this system.184A popular way of assessing thermodynamic data for a series of related compounds is using enthalpy–entropy compensation plots. The enthalpy–entropy compensation phenomenon appears to occur in many, if not all, binding studies and is demonstrated by a linear correlation between the enthalpic and entropic contributions to binding for a series of perturbations or changes in experimental variable, such as temperature, series of homologous compounds or mutants.186 Many dispute the significance of enthalpy–entropy compensation, claiming it demonstrates the known thermodynamic laws and the limitations of the values measured.186,187 Compensation between enthalpy and entropy terms is also likely to be evident where the range of ΔG°a values measured is relatively small; this limited range is likely to occur both from the range of values measurable using techniques such as ITC and the values imposed by biology.186,188–190 In addition the intrinsic properties of water are also likely to be important in the compensation. Upon ligand binding a favourable enthalpic contribution suggests solvent molecules are involved in strong hydrogen bonding interactions, but this is likely to be associated with an entropic cost due to the small configurational freedom in the system, whereas conversely the loss of bound water molecules contributes favourably to entropy and the disorder of the system, but unfavourably to the enthalpy term as hydrogen bonding interactions are lost.187,191–193
An enthalpy–entropy plot for the thermodynamic data determined for the 18 inhibitors with a β-glucosidase shows a line of best fit with a slope of 0.93 and correlation of 0.91 (Fig. 5b).184 No related inhibitors, however, such as those which may mimic the charge at the transition state or those which may mimic the geometry, are closely grouped in the compensation plot. In fact very closely related compounds which only differ by one atom type were observed at opposite ends of the trend. The ITC data for three other inhibitors tested with the β-glucosidase since the original publication, two 1-deoxy-6-oxa-N-(thiocarbamoyl)calystegines194 and a 3-imino-2-thia-(+)-castanospermine analogue195 (shown in Fig. 5b), also lie on the best fit line through the points. The castanospermine analogue was shown to differ from the other compounds in that it bound with an unfavourable enthalpic contribution which was more than compensated by a large entropy; despite this different thermodynamic signature, this point remarkably still lies on the line. It is interesting to note that a study investigating putative transition state analogues with the human purine nucleoside phosphorylase, some of which have picomolar affinity, also show strong enthalpy–entropy compensation. In this plot, however, related compounds did tend to cluster in one region of the plot.196
The differences in the (closely) related inhibitors studied are most likely attributed to the effects of solvation and desolvation. Whilst ITC studies can quantify the thermodynamics of inhibitor binding to protein and the subsequent displacement of water molecules during this process, nothing is known about the solvation state of either the protein or inhibitor alone. Homans and co-workers have demonstrated that desolvation of the inhibitor upon binding is likely to contribute significantly;197 it may, therefore, only be when more sophisticated methods are available to measure or model these important contributions that the true thermodynamics of inhibitor binding are revealed.
Slow onset inhibition
Slow onset inhibition, a phenomenon characterised by an initial rapid catalytic rate by an enzyme in the presence of an inhibitor, followed by a decrease to a slower steady state rate after an amount of time (Fig. 5c), has been observed with a number of glycosidase inhibitors, including nojirimycin 1,198,199 deoxynojirimycin 2,199 acarbose 21,199 isofagomine 9 and analogues,101,184,200,201 castanospermine 5,76,184 noeuromycin 10,184 azafagomine 12,184 hydroximolactam 14,184 glucotetrazole 15,184 glucoimidazole 16184 and derivatives such as 17125,184 and isofagomine lactam 18.184 Various reasons have been offered as to why inhibitors may display slow onset. The initial rapid reaction rate may represent formation of the enzyme-inhibitor complex, after which isomerisation55,202–204 or conformational changes76,203,205,206 of the inhibitor or protein causes formation of a more potent inhibitory species. It has also been suggested the lag phase for inhibition may allow a time-dependent protonation of the enzyme or inhibitor.203,206,207 Slow onset inhibition may also be a consequence of a slow interaction (less than the rate of diffusion) between the inhibitor and enzyme.202,203,208Schramm and co-workers have suggested that the presence of slow onset inhibition is a criterion for an inhibitor to be a true transition state mimic (although not exclusively).178,209 They believe that slow onset is evident among transition state mimicking inhibitors because they bind to an enzyme that is conformationally optimised to bind the substrate in the ground state. The lag phase before the true inhibitory potential of an inhibitor becomes apparent is therefore attributed to conformational changes that must occur in the active site, which will be less efficient with a transition state analogue than with the ground state substrate for which it optimised.55,178,209 Although evidence of slow onset inhibition is a useful guide when studying inhibitors that are putative transition state mimics, it is highly likely that inhibitors which do not comply also display the phenomenon; this is strongly suggested merely by the number of glycosidase inhibitors which have been reported to show it, as detailed in the previous paragraph. It may also be possible that all inhibitors show slow onset inhibition to some degree, but is more apparent when measuring high affinity inhibition constants at low inhibitor concentration.55
pH dependence of inhibition
Another proposed criterion for analysing which inhibitors are likely to be true transition state mimics is the comparison of the profiles for pH dependence of inhibition and catalysis.210 A difference in the pH dependence for catalysis vs. inhibition, where, for example, an inhibitor inhibits most effectively when the enzyme is largely inactive, suggests it cannot be a true transition state mimic, as, by definition, a catalytically inactive enzyme cannot bind the transition state.211,212 A study which examined the pH profiles for both catalysis and inhibition using 18 putative transition state analogues indicated that in very few cases did the profile for inhibition mirror that for catalysis.184 This was only apparent for tetrahydrooxazine (11), the hydroximolactam analogues (14) (Fig. 5d) and glucotetrazole (15); each of these compounds has a low pKa value (3.6 for 11,104 4.8 for 14111 and −4.0 for 15124) and so the titration is likely to reflect the catalytic residues rather than the inhibitor itself. In fact, it has been suggested the pH profile for inhibition will only mirror catalysis when the pKa of the inhibitor is below that of the acidic catalytic residue of the enzyme,213 but if this was the case it would seem intuitively incorrect to assume these were transition state analogues. A counter-argument can be made here though, as at the transition state both the nucleophile and acid/base residues may both be partially deionised, and thus tight-binding displayed by inhibitors at high pH where the enzyme is doubly deionised may actually better reflect the real situation.213 If this was the case then deoxynojirimycin 2, castanospermine 5, calystegine B28, isofagomine 9, noeuromycin 10, glucoimidazole 16 and some of it derivatives, such as 17, would all be candidates as transition state analogues.It should be acknowledged, however, that analysis of pH profiles for catalysis and inhibition are extremely circumspect. McIntosh et al. have described how the pKa values of the catalytic residues of a xylanase ‘cycle’ during catalysis, with changes of up to 2.5 pH units.214 Although it is assumed in most cases that the acidic limb of pH dependence for catalysis demonstrates titration of the nucleophile residue and the basic limb titration of the acid/base residue, and that at the pH optimum for catalysis, the acid/base will be protonated and the nucleophile deprotonated, it is impossible to assign this unequivocally in each case. Measurement of the microscopic pKa values of the acid/base and nucleophile demonstrate that they are heavily reliant on the ionisation state of the other.214 If the pKa values of the catalytic residues change by as much as 2.5 pH units during hydrolysis, the question has to be raised as to their likely values at the transition state, but this can never be determined. It does have wide implications, however, for the likely pKa values of the catalytic residues when bound to inhibitors that may mimic the transition state. The presence of inhibitors complicate the matter even further as they themselves may be protonated, or indeed their protonation state may change upon binding to the enzyme. Atomic resolution X-ray crystallography has indicated that upon inhibitor binding the ionisation state of the catalytic residues can easily change (although they themselves may be influenced by the crystallisation conditions); for example, the xylobio-derived forms of 2215 and 18137 bound to an enzyme where the nucleophile and acid/base had reversed protonation states than required for catalysis, and the cellobio-derived form of 9 bound to an enzyme where both residues were deionised.101 Catalytic residues and inhibitors may also be subject to pKa changes upon binding, meaning the pKa values determined for each in the free state are meaningless.213 Both Knowles216 and Brocklehurst217 have warned about the risks involved with analysing pH dependence data, both for catalysis and inhibition. The pH profiles for inhibition are likely to reflect composites of the free enzyme, free inhibitor and the enzyme-inhibitor complex.216
Conclusions
This article, and a number of other reviews in the glycosidase inhibition field (including Ref. 64–70), have demonstrated that the quest for potent and selective inhibitors is extremely active at present. The involvement of glycosidases in lysosomal storage disorders, cancer, viral infections and diabetes demonstrates the need to continue strong and fruitful research in this area. It is, however, perhaps a little alarming to acknowledge the small gains in potency made in these inhibitors over the past 50 years or so. As Wolfenden has demonstrated the binding potential of the transition state to be approximately 10−22 M,56 it has been proposed that if features of the transition state could be included in the design of inhibitors, then higher potency should result. In many cases this has been achieved on an intuitive level, where aspects of the transition state charge or geometry have been introduced into inhibitors. However, the exact nature of the transition state is unknown, and indeed will differ between individual enzymes, and so more quantitative methods are required to gain confidence in the transition state analogy. A number of criteria for transition state mimicry have been proposed, and have been discussed in relation to inhibitors where they have been tested. Of these, it is perhaps the LFERs that use the most quantitative means to test the likelihood of an inhibitor mimicking the transition state, although still gives a qualitative answer. It is interesting also to note that different conclusions were drawn from the same ‘classes’ of inhibitors with the different enzymes described here which may reflect the subtleties of the transition state in each case. This point has been highlighted previously by Schramm and colleagues by demonstrating that the bovine, human and Mycobacterium species forms of an enzyme utilised either an early (more like the substrate) or late (more like the product) transition state, and that by incorporating features into inhibitors to mimic each of these structures, different compounds inhibited each of the enzymes optimally.60,178 Indeed, it has also been suggested that the structure of the transition state may change with the evolution of an enzyme.218 It is, of course, not possible for any compound to mimic the transition state in its entirety as it would have to include partially formed and broken bonds, a partial positive charge and a trigonal centre which is chemically challenging; harnessing the full binding potential of the transition state is therefore never going to be realised.It can, of course, be argued that highly effective inhibitors do not have to mimic the transition state and there are a plethora of potent and specific drugs in clinical use as examples. A favourable combination of the necessary hydrogen bond, electrostatic and hydrophobic interactions can lead to inhibitors that bear little resemblance to the transition state. A picomolar inhibitor (methotrexate) of dihydrofolate reductase was observed using X-ray crystallography to actually bind upside down when compared to the substrate, even though very similar interactions were made, demonstrating that Nature can be unpredictable.219 Indeed an array of glycosidase inhibitors have been identified which not only lack features of the transition state, but any resemblance of a carbohydrate moiety; these include inhibitory proteins,220,221 cyclopeptides,222 amines (which is why Tris buffer is often observed to inhibit glycosidases)211 and phenyl-imidazole compounds.223
Although the potency of glycosidase inhibitors has been slow to improve over the past half century, it is perhaps only in the past decade or so that the detailed understanding of reaction mechanism and transition state structure, chemical synthetic methods to allow inhibitors incorporating different features to be synthesised and computer modelling to aid inhibitor design, in addition to techniques such as kinetics for measurements of LFERs and ITC for determination of thermodynamic signatures, have developed sufficiently to realise their full potential in inhibitor design. Schramm and colleagues have elegantly demonstrated the power of combining techniques such as X-ray crystallography, computer modelling and kinetic isotope effects to deduce the structure of the transition state and design analogues,62,63,178,209 which has led to the production of picomolar or femtomolar inhibitors of enzymes with a number of medical applications. Their studies have highlighted some of the subtleties of transition state mimic design, which, in conjunction with the enhanced methodologies should now be used to catalyse the advancement of potency for glycosidase inhibitors. Given the extremely tight binding of the transition state in glycoside hydrolysis, the design of inhibitors that mimic its structure must be one of the most efficient ways of achieving highly potent drugs in the quest for inhibiting glycosidases in the future.
Acknowledgements
T.M.G is a Sir Henry Wellcome postdoctoral fellow and a Michael Smith for Health Research trainee award holder, and G.J.D is a Royal Society Wolfson Research Merit Award recipient.Notes and references
- G. J. Davies, T. M. Gloster and B. Henrissat, Curr. Opin. Struct. Biol., 2005, 15, 637–645 CrossRef CAS.
- R. A. Laine, Glycobiology, 1994, 4, 759–767 CAS.
- R. Wolfenden, X. Lu and G. Young, J. Am. Chem. Soc., 1998, 120, 6814–6815 CrossRef CAS.
- Y. Suzuki, S. Ichinomiya, M. Kurosawa, M. Ohkubo, H. Watanabe, H. Iwasaki, J. Matsuda, Y. Noguchi, K. Takimoto, M. Itoh, M. Tabe, M. Iida, T. Kubo, S. Ogawa, E. Nanbe, K. Higaki, K. Ohno and R. O. Brady, Ann. Neurol., 2007, 62, 671–675 CrossRef CAS.
- A. R. Sawkar, W.-C. Cheng, E. Beutler, C.-H. Wong, W. E. Balch and J. W. Kelly, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 15428–15433 CrossRef CAS.
- G. H.-F. Yam, N. Bosshard, C. Zuber, B. Steinmann and J. Roth, Am. J. Physiol.: Cell Physiol., 2006, 290, C1076–C1082 CAS.
- J.-Q. Fan, S. Ishii, N. Asano and Y. Suzuki, Nat. Med., 1999, 5, 112–115 CrossRef CAS.
- A. J. Krentz and C. J. Bailey, Drugs, 2005, 65, 385–411 CrossRef CAS.
- B. Göke and C. Herrmann-Rinke, Diabetes Metab. Rev., 1998, 14, S31–S38 CrossRef CAS.
- A. Mehta, N. Zitzmann, P. M. Rudd, T. M. Block and R. A. Dwek, FEBS Lett., 1998, 430, 17–22 CrossRef CAS.
- M. von Itzstein, Nat. Rev. Drug Discovery, 2007, 6, 967–974 CrossRef CAS.
- J. Zhang and W. Xu, Mini-Rev. Med. Chem., 2006, 6, 428–448.
- M.-J. Papandréou, R. Barbouche, R. Guieu, M. P. Kieny and E. Fenouillet, Mol. Pharmacol., 2002, 61, 186–193 CrossRef CAS.
- D. Sailer and G. Röder, Arzneimittelforschung, 1980, 30, 2182–2185 CAS.
- N. Katsilambros, P. Philippides, T. Toskas, J. Protopapas, D. Frangaki, M. Marangos, P. Siskoudis, K. Anastasopoulou, H. Xefteri and I. Hillebrand, Arzneimittelforschung, 1986, 36, 1136–1138 CAS.
- M. von Itzstein, W.-Y. Wu, G. B. Kok, M. S. Pegg, J. C. Dyason, B. Jin, T. V. Phan, M. L. Smythe, H. F. White, S. W. Oliver, P. M. Colman, J. N. Varghese, D. M. Ryan, J. M. Woods, R. C. Bethell, V. J. Hotham, J. M. Cameron and C. R. Penn, Nature, 1993, 363, 418–423 CrossRef CAS.
- F. G. Hayden, J. J. Treanor, R. F. Betts, M. Lobo, J. D. Esinhart and E. K. Hussey, J. Am. Med. Assoc., 1996, 275, 295–299 CrossRef CAS.
- C. U. Kim, W. Lew, M. A. Williams, H. Liu, L. Zhang, S. Swaminathan, N. Bischofberger, M. S. Chen, D. B. Mendel, C. Y. Tai, W. G. Laver and R. C. Stevens, J. Am. Chem. Soc., 1997, 119, 681–690 CrossRef CAS.
- F. G. Hayden, R. L. Atmar, M. Schilling, C. Johnson, D. Poretz, D. Paar, L. Huson, P. Ward and R. G. Mills, N. Engl. J. Med., 1999, 341, 1336–1343 CrossRef CAS.
- S. A. Yuzwa, M. S. Macauley, J. E. Heinonen, X. Shan, R. J. Dennis, Y. He, G. E. Whitworth, K. A. Stubbs, E. J. McEachern, G. J. Davies and D. J. Vocadlo, Nat. Chem. Biol., 2008, 4, 483–490 CrossRef CAS.
- M. S. Macauley, A. K. Bubb, C. Martinez-Fleites, G. J. Davies and D. J. Vocadlo, J. Biol. Chem., 2008, 283, 34687–34695 CrossRef CAS.
- B. L. Cantarel, P. M. Coutinho, C. Rancurel, T. Bernard, V. Lombard and B. Henrissat, Nucleic Acids Res., 2008, 37, D233–238.
- B. Henrissat and G. Davies, Curr. Opin. Struct. Biol., 1997, 7, 637–644 CrossRef CAS.
- V. L. Y. Yip, A. Varrot, G. J. Davies, S. S. Rajan, X. Yang, J. Thompson, W. F. Anderson and S. G. Withers, J. Am. Chem. Soc., 2004, 126, 8354–8355 CrossRef CAS.
- Q. P. Liu, G. Sulzenbacher, H. Yuan, E. P. Bennett, G. Pietz, K. Saunders, J. Spence, E. Nudelman, S. B. Levery, T. White, J. M. Neveu, W. S. Lane, Y. Bourne, M. L. Olsson, B. Henrissat and H. Clausen, Nat. Biotechnol., 2007, 25, 454–464 CrossRef CAS.
- T. M. Gloster, J. P. Turkenburg, J. R. Potts, B. Henrissat and G. J. Davies, Chem. Biol., 2008, 15, 1058–1067 CrossRef CAS.
- M. Kitamura, M. Okuyama, F. Tanzawa, H. Mori, Y. Kitago, H. Watanabe, A. Kimura, I. Tanaka and M. Yao, J. Biol. Chem., 2008, 283, 36328–36337 CrossRef CAS.
- R. Kuroki, L. H. Weaver and B. W. Matthews, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 8949–8954 CrossRef CAS.
- A. M. Thunnissen, H. J. Rozeboom, K. H. Kalk and B. W. Dijkstra, Biochemistry, 1995, 34, 12729–12737 CrossRef CAS.
- D. E. Koshland, Jr, Biol. Rev., 1953, 28, 416–436 CAS.
- A. Vasella, G. J. Davies and M. Böhm, Curr. Opin. Chem. Biol., 2002, 6, 619–629 CrossRef CAS.
- D. J. Vocadlo and G. J. Davies, Curr. Opin. Chem. Biol., 2008, 12, 539–555 CrossRef CAS.
- C. S. Rye and S. G. Withers, Curr. Opin. Chem. Biol., 2000, 4, 573–580 CrossRef CAS.
- G. J. Davies, M. L. Sinnott, and S. G. Withers, in Comprehensive biological catalysis. A mechanistic reference, ed. M. L. Sinnott, Academic Press, London, 1998, Volume I, pp. 119–209 Search PubMed.
- M. L. Sinnott, Chem. Rev., 1990, 90, 1171–1202 CrossRef.
- A. C. Terwisscha van Scheltinga, S. Armand, K. H. Kalk, A. Isogai, B. Henrissat and B. Dijkstra, Biochemistry, 1995, 34, 15619–15623 CrossRef.
- I. Tews, A. Perrakis, A. Oppenheim, Z. Dauter, K. S. Wilson and C. E. Vorgias, Nat. Struct. Biol., 1996, 3, 638–648 CrossRef CAS.
- Z. Markovic-Housley, G. Miglierini, L. Soldatova, P. J. Rizkallah, U. Müller and T. Schirmer, Structure, 2000, 8, 1025–1035 CrossRef CAS.
- S. Knapp, D. Vocadlo, Z. Gao, B. Kirk, J. Lou and S. G. Withers, J. Am. Chem. Soc., 1996, 118, 6804–6805 CrossRef CAS.
- M. S. Macauley, G. E. Whitworth, A. W. Debowski, D. Chin and D. J. Vocadlo, J. Biol. Chem., 2005, 280, 25313–25322 CrossRef CAS.
- M. Umekawa, W. Huang, B. Li, K. Fujita, H. Ashida, L. Wang and K. Yamamoto, J. Biol. Chem., 2008, 283, 4469–4479 CAS.
- D. W. Abbott, M. S. Macauley, D. J. Vocadlo and A. B. Boraston, J. Biol. Chem., 2009, 284, 11676–11689 CAS.
- C. Martinez-Fleites, J. E. Korczynska, G. J. Davies, M. J. Cope, J. P. Turkenburg and E. J. Taylor, Carbohydr. Res., 2009, 344, 1753–1757 CrossRef CAS.
- A. G. Watts, I. Damager, M. L. Amaya, A. Buschiazzo, P. Alzari, A. C. Frasch and S. G. Withers, J. Am. Chem. Soc., 2003, 125, 7532–7533 CrossRef CAS.
- I. Damager, S. Buchini, M. F. Amaya, A. Buschiazzo, P. Alzari, A. C. Frasch, A. G. Watts and S. G. Withers, Biochemistry, 2008, 47, 3507–3512 CrossRef CAS.
- K. Stummeyer, A. Dickmanns, M. Muhlenhoff, R. Gerardy-Schahn and R. Ficner, Nat. Struct. Mol. Biol., 2005, 12, 90–96 CrossRef CAS.
- T. J. Morley, L. M. Willis, C. Whitfield, W. W. Wakarchuk and S. G. Withers, J. Biol. Chem., 2009, 284, 17404–17410 CrossRef CAS.
- B. P. Rempel and S. G. Withers, Glycobiology, 2008, 18, 570–586 CrossRef CAS.
- R. Wolfenden and M. J. Snider, Acc. Chem. Res., 2001, 34, 938–945 CrossRef CAS.
- L. Pauling, Chem. Eng. News, 1946, 24, 1375–1377 CAS.
- L. Pauling, Nature, 1948, 161, 707–709 CrossRef CAS.
- G. J. Davies, V. M.-A. Ducros, A. Varrot and D. L. Zechel, Biochem. Soc. Trans., 2003, 31, 523–527 CAS.
- J. O'Connor Westerik and R. Wolfenden, J. Biol. Chem., 1972, 247, 8195–8197.
- R. C. Thompson, Biochemistry, 1973, 12, 47–51 CrossRef CAS.
- M. M. Mader and P. A. Bartlett, Chem. Rev., 1997, 97, 1281–1301 CrossRef CAS.
- R. Wolfenden, M. Snider, C. Ridgway and B. Miller, J. Am. Chem. Soc., 1999, 121, 7419–7420 CrossRef CAS.
- M. Snider, S. Gaunitz, C. Ridgway, S. A. Short and R. Wolfenden, Biochemistry, 2000, 39, 9746–9753 CrossRef CAS.
- V. L. Schramm, J. Biol. Chem., 2007, 282, 28297–28300 CrossRef CAS.
- G. A. Kicska, L. Long, H. Hörig, C. Fairchild, P. C. Tyler, R. H. Furneaux, V. L. Schramm and H. L. Kaufman, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 4593–4598 CrossRef CAS.
- A. Lewandowicz, E. A. Ringia, L. M. Ting, K. Kim, P. C. Tyler, G. B. Evans, O. V. Zubkova, S. Mee, G. F. Painter, D. H. Lenz, R. H. Furneaux and V. L. Schramm, J. Biol. Chem., 2005, 280, 30320–30328 CrossRef CAS.
- K. Balakrishnan, R. Nimmanapalli, F. Ravandi, M. J. Keating and V. Gandhi, Blood, 2006, 108, 2392–2398 CrossRef CAS.
- J. E. Lee, V. Singh, G. B. Evans, P. C. Tyler, R. H. Furneaux, K. A. Cornell, M. K. Riscoe, V. L. Schramm and P. L. Howell, J. Biol. Chem., 2005, 280, 18274–18282 CAS.
- V. Singh, G. B. Evans, D. H. Lenz, J. M. Mason, K. Clinch, S. Mee, G. F. Painter, P. C. Tyler, R. H. Furneaux, J. E. Lee, P. L. Howell and V. L. Schramm, J. Biol. Chem., 2005, 280, 18265–18273 CAS.
- V. H. Lillelund, H. H. Jensen, X. Liang and M. Bols, Chem. Rev., 2002, 102, 515–553 CrossRef CAS.
- A. E. Stütz, ed., Iminosugars as glycosidase inhibitors. Nojirimycin and beyond, Wiley-VCH, Weinheim, 1999 Search PubMed.
- N. Asano, R. J. Nash, R. J. Molyneux and G. W. J. Fleet, Tetrahedron: Asymmetry, 2000, 11, 1645–1680 CrossRef CAS.
- T. D. Heightman and A. T. Vasella, Angew. Chem., Int. Ed., 1999, 38, 750–770 CrossRef CAS.
- A. A. Watson, G. W. J. Fleet, N. Asano, R. J. Molyneux and R. J. Nash, Phytochemistry, 2001, 56, 265–295 CrossRef CAS.
- N. Asano, Glycobiology, 2003, 13, 93R–104R CrossRef CAS.
- N. Asano, Cell Mol. Life Sci., 2009, 66, 1479–1492 CrossRef CAS.
- S. Inouye, T. Tsuroka and T. Nida, J. Antibiot., 1966, 19, 288–292 CAS.
- S. Inouye, T. Tsuruoka, T. Ito and T. Niida, Tetrahedron, 1968, 24, 2124–2144.
- H. Paulsen, I. Sangster and K. Heyns, Chem. Ber., 1967, 100, 802–815.
- M. Yagi, T. Kouno, Y. Aoyagi and H. Murai, Nippon Nogei Kagaku Kaishi, 1976, 50, 571–572 Search PubMed.
- D. D. Schmidt, W. Frommer, L. Müller and E. Truscheit, Naturwissenschaften, 1979, 66, 584–585 CAS.
- G. Legler, Adv. Carbohydr. Chem. Biochem., 1990, 48, 319–384 CAS.
- M. Koyama and S. Sakamura, Agric. Biol. Chem., 1974, 38, 1111–1112 CAS.
- A. Kato, N. Asano, H. Kizu, K. Matsui, A. A. Watson and R. J. Nash, J. Nat. Prod., 1997, 60, 312–314 CrossRef CAS.
- F. M. Platt, G. R. Neises, R. A. Dwek and T. D. Butters, J. Biol. Chem., 1994, 269, 8362–8365 CAS.
- A. Zimran and D. Elstein, Philos. Trans. R. Soc. London, Ser. B, 2003, 358, 961–966 CrossRef CAS.
- L. D. Hohenschutz, E. A. Bell, P. J. Jewess, D. P. Leworthy, R. J. Pryce, E. Arnold and J. Clardy, Phytochemistry, 1981, 20, 811–814 CrossRef CAS.
- J.-L. Reymond, A. A. Pinkerton and P. Vogel, J. Org. Chem., 1991, 56, 2128–2135 CrossRef CAS.
- K. Whitby, T. C. Pierson, B. Geiss, K. Lane, M. Engle, Y. Zhou, R. W. Doms and M. S. Diamond, J. Virol., 2005, 79, 8698–8706 CrossRef CAS.
- S. M. Colegate, P. R. Dorling and C. R. Huxtable, Aust. J. Chem., 1979, 32, 2257–2264 CAS.
- G. W. J. Fleet, M. J. Gough and P. W. Smith, Tetrahedron Lett., 1984, 25, 1853–1856 CrossRef CAS.
- H. Kayakiri, S. Takase, T. Shibata, M. Okamoto, H. Terano and M. Hashimoto, J. Org. Chem., 1989, 54, 4015–4016 CrossRef CAS.
- H. Kayakiri, C. Kasahara, T. Oku and M. Hashimoto, Tetrahedron Lett., 1990, 31, 225–226 CrossRef CAS.
- A. D. Elbein, J. E. Tropea, M. Mitchell and G. P. Kaushal, J. Biol. Chem., 1990, 265, 15599–15605 CAS.
- A. D. Elbein, R. Solf, P. R. Dorling and K. Vosbeck, Proc. Natl. Acad. Sci. U. S. A., 1981, 78, 7393–7397 CrossRef CAS.
- P. E. Goss, M. A. Baker, J. P. Carver and J. W. Dennis, Clin. Cancer Res., 1995, 1, 935–944 CAS.
- D. Tepfer, A. Goldmann, N. Pamboukdjian, M. Maille, A. Lepingle, D. Chevalier, J. Dénarié and C. Rosenberg, J. Bacteriol., 1988, 170, 1153–1161 CAS.
- B. Dräger, Nat. Prod. Rep., 2004, 21, 211–223 RSC.
- P. R. Skaanderup and R. Madsen, J. Org. Chem., 2003, 68, 2115–2122 CrossRef CAS.
- T. M. Gloster, R. Madsen and G. J. Davies, ChemBioChem, 2006, 7, 738–742 CrossRef CAS.
- T. M. Jespersen and M. Bols, Tetrahedron, 1994, 50, 13449–13460 CrossRef CAS.
- T. M. Jespersen, W. Dong, M. R. Sierks, T. Skrydstrup, I. Lundt and M. Bols, Angew. Chem., Int. Ed. Engl., 1994, 33, 1778–1779 CrossRef.
- J. Andersch and M. Bols, Chem.–Eur. J., 2001, 7, 3744–3747 CrossRef CAS.
- Y. Ichikawa and Y. Igarashi, Tetrahedron Lett., 1995, 36, 4585–4586 CrossRef CAS.
- Y. Ichikawa, Y. Igarashi, M. Ichikawa and Y. Suhara, J. Am. Chem. Soc., 1998, 120, 3007–3018 CrossRef CAS.
- Y. J. Kim, M. Ichikawa and Y. Ichikawa, J. Org. Chem., 2000, 65, 2599–2602 CrossRef CAS.
- A. Varrot, C. A. Tarling, J. M. Macdonald, R. V. Stick, D. L. Zechel, S. G. Withers and G. J. Davies, J. Am. Chem. Soc., 2003, 125, 7496–7497 CrossRef CAS.
- M. N. Namchuk, J. D. McCarter, A. Becalski, T. Andrews and S. G. Withers, J. Am. Chem. Soc., 2000, 122, 1270–1277 CrossRef CAS.
- H. Z. Liu, X. F. Liang, H. Søhoel, A. Bülow and M. Bols, J. Am. Chem. Soc., 2001, 123, 5116–5117 CrossRef CAS.
- P. Bach and M. Bols, Tetrahedron Lett., 1999, 40, 3461–3464 CrossRef CAS.
- M. Bols, R. G. Hazell and I. B. Thomsen, Chem.–Eur. J., 1997, 3, 940–947 CAS.
- X. Liang and M. Bols, J. Org. Chem., 1999, 64, 8485–8488 CrossRef CAS.
- B. V. Ernholt, I. B. Thomsen, A. Lohse, I. W. Plesner, K. B. Jensen, R. G. Hazell, X. Liang, A. Jakobsen and M. Bols, Chem.–Eur. J., 2000, 6, 278–287 CrossRef CAS.
- M. K. Tong, G. Papandreou and B. Ganem, J. Am. Chem. Soc., 1990, 112, 6137–6139 CrossRef CAS.
- B. Ganem and G. Papandreou, J. Am. Chem. Soc., 1991, 113, 8984–8985 CrossRef CAS.
- G. Papandreou, M. K. Tong and B. Ganem, J. Am. Chem. Soc., 1993, 115, 11682–11690 CrossRef.
- R. Hoos, A. Vasella, K. Rupitz and S. G. Withers, Carbohydr. Res., 1997, 298, 291–298 CrossRef CAS.
- Y. Blériot, A. Genre-Grandpierre and C. Tellier, Tetrahedron Lett., 1994, 35, 1867–1870 CrossRef CAS.
- Y. Blériot, T. Dintinger, A. Genre-Grandpierre, M. Padrines and C. Tellier, Bioorg. Med. Chem. Lett., 1995, 5, 2655–2660 CrossRef CAS.
- M.-P. Heck, S. P. Vincent, B. W. Murray, F. Bellamy, C.-H. Wong and C. Mioskowski, J. Am. Chem. Soc., 2004, 126, 1971–1979 CrossRef CAS.
- R. Hoos, A. B. Naughton, W. Thiel, A. Vasella, W. Weber, K. Rupitz and S. G. Withers, Helv. Chim. Acta, 1993, 76, 2666–2686 CrossRef CAS.
- S. Vonhoff, T. D. Heightman and A. Vasella, Helv. Chim. Acta, 1998, 81, 1710–1725 CrossRef CAS.
- S. Vonhoff, K. Piens, M. Pipelier, C. Braet, M. Claeyssens and A. Vasella, Helv. Chim. Acta, 1999, 82, 963–980 CrossRef CAS.
- P. Ermert and A. Vasella, Helv. Chim. Acta, 1991, 74, 2043–2053 CAS.
- P. Ermert, A. Vasella, M. Weber, K. Rupitz and S. G. Withers, Carbohydr. Res., 1993, 250, 113–128 CrossRef CAS.
- T. D. Heightman, P. Ermert, D. Klein and A. Vasella, Helv. Chim. Acta, 1995, 78, 514–532 CrossRef CAS.
- T. Granier, N. Panday and A. Vasella, Helv. Chim. Acta, 1997, 80, 979–987 CrossRef CAS.
- J. Pabba, N. Mohal and A. Vasella, Helv. Chim. Acta, 2006, 89, 1373–1386 CrossRef CAS.
- N. Panday, T. Granier and A. Vasella, Helv. Chim. Acta, 1998, 81, 475–490 CrossRef CAS.
- N. Panday and A. Vasella, Synthesis, 1999, SI, 1459–1468 CrossRef.
- N. Panday, Y. Canac and A. Vasella, Helv. Chim. Acta, 2000, 83, 58–79 CrossRef CAS.
- B. Shanmugasundaram and A. Vasella, Helv. Chim. Acta, 2005, 88, 2593–2602 CrossRef CAS.
- M. Terinek and A. Vasella, Helv. Chim. Acta, 2004, 87, 3035–3049 CrossRef CAS.
- M. Terinek and A. Vasella, Helv. Chim. Acta, 2005, 88, 10–22 CrossRef CAS.
- M. Terinek and A. Vasella, Helv. Chim. Acta, 2003, 86, 3482–3509 CrossRef CAS.
- T. D. Heightman, M. Locatelli and A. Vasella, Helv. Chim. Acta, 1996, 79, 2190–2200 CrossRef CAS.
- G. J. Davies, K. S. Wilson and B. Henrissat, Biochem. J., 1997, 321, 557–559 CAS.
- M. Hrmova, R. de Gori, B. J. Smith, A. Vasella, J. N. Vargese and G. B. Fincher, J. Biol. Chem., 2004, 279, 4970–4980 CAS.
- M. Hrmova, V. A. Streltsov, B. J. Smith, A. Vasella, J. N. Vargese and G. B. Fincher, Biochemistry, 2005, 44, 16529–16539 CrossRef CAS.
- L. E. Tailford, W. A. Offen, N. L. Smith, C. Dumon, C. Morland, J. Gratien, M. P. Heck, R. V. Stick, Y. Bleriot, A. Vasella, H. J. Gilbert and G. J. Davies, Nat. Chem. Biol., 2008, 4, 306–312 CrossRef CAS.
- T. M. Gloster, S. Roberts, G. Perugino, M. Rossi, M. Moracci, N. Panday, M. Terinek, A. Vasella and G. J. Davies, Biochemistry, 2006, 45, 11879–11884 CrossRef CAS.
- S. J. Williams, V. Notenboom, J. Wicki, D. R. Rose and S. G. Withers, J. Am. Chem. Soc., 2000, 122, 4229–4230 CrossRef CAS.
- T. Gloster, S. J. Williams, C. A. Tarling, S. Roberts, C. Dupont, P. Jodoin, F. Shareck, S. G. Withers and G. J. Davies, Chem. Commun., 2003, 944–945 RSC.
- V. H. Lillelund, H. Liu, X. Liang, H. Søhoel and M. Bols, Org. Biomol. Chem., 2003, 1, 282–287 RSC.
- X. Chen, Y. Fan, Y. Zheng and Y. Shen, Chem. Rev., 2003, 103, 1955–1977 CrossRef CAS.
- Y. Kameda and S. Horii, J. Chem. Soc., Chem. Commun., 1972, 746–747 RSC.
- H. Paulsen and F. R. Heiker, Angew. Chem., Int. Ed., 1980, 18, 904–905 CrossRef.
- S. Ogawa, T. Ogawa, Y. Iasawa, T. Toyokuni, N. Chida and T. Suami, J. Org. Chem., 1984, 49, 2594–2599 CrossRef CAS.
- Y. Kameda, N. Asano, T. Yamaguchi and K. Matsui, J. Antibiot., 1987, 40, 563–565.
- R. P. Gibson, T. M. Gloster, S. Roberts, R. A. Warren, I. Storch de Gracia, A. Garcia, J. L. Chiara and G. J. Davies, Angew. Chem., Int. Ed., 2007, 46, 4115–4119 CrossRef CAS.
- A. E. Aleshin, B. Stoffer, L. M. Firsov, B. Svensson and R. B. Honzatko, Biochemistry, 1996, 35, 8319–8328 CrossRef CAS.
- S. Ezaki, J. Biochem., 1940, 32, 87–90.
- Hobikoshi, J. Biochem., 1942, 35, 39–59.
- D. H. Leaback, Biochem. Biophys. Res. Commun., 1968, 32, 1025–1030 CrossRef CAS.
- W. P. Burmeister, S. Cottaz, P. Rollin, A. Vasella and B. Henrissat, J. Biol. Chem., 2000, 275, 39385–39393 CrossRef CAS.
- D. H. Juers, T. D. Heightman, A. Vasella, J. D. McCarter, L. Mackenzie, S. G. Withers and B. W. Matthews, Biochemistry, 2001, 40, 14781–14794 CrossRef CAS.
- E. T. Reese, F. W. Parrish and M. Ettlinger, Carbohydr. Res., 1971, 18, 381–388 CrossRef CAS.
- C. L. Combes and G. G. Birch, Food Chem., 1988, 27, 283–298 CrossRef CAS.
- T. Kajimoto, K. K.-C. Liu, R. L. Pederson, Z. Zhong, Y. Ichikawa, J. A. Porco and C.-H. Wong, J. Am. Chem. Soc., 1991, 113, 6187–6196 CrossRef CAS.
- Y. Le Merrer, M. Fuzier, I. Dosbaa, M.-J. Foglietti and J. C. Depezay, Tetrahedron, 1997, 53, 16731–16746 CrossRef CAS.
- M. Yoshikawa, T. Murakami, H. Shimada, H. Matsuda, J. Yamahara, G. Tanabe and O. Muraoka, Tetrahedron Lett., 1997, 38, 8367–8370 CrossRef CAS.
- H. Yuasa, J. Takada and H. Hashimoto, Tetrahedron Lett., 2000, 41, 6615–6618 CrossRef CAS.
- A. Ghavami, B. D. Johnston and B. M. Pinto, J. Org. Chem., 2001, 66, 2312–2317 CrossRef CAS.
- A. Ghavami, B. D. Johnston, M. T. Jensen, B. Svensson and B. M. Pinto, J. Am. Chem. Soc., 2001, 123, 6268–6271 CrossRef CAS.
- S. Mohan and B. M. Pinto, Carbohydr. Res., 2007, 342, 1551–1580 CrossRef CAS.
- L. Svansson, B. D. Johnston, J. H. Gu, B. Patrick and B. M. Pinto, J. Am. Chem. Soc., 2000, 122, 10769–10775 CrossRef CAS.
- N. S. Kumar and B. M. Pinto, J. Org. Chem., 2006, 71, 1262–1264 CrossRef CAS.
- M. Horsch, L. Hoesch, A. Vasella and D. M. Rast, Eur. J. Biochem., 1991, 197, 815–818 CAS.
- F. V. Rao, H. C. Dorfmueller, F. Villa, M. Allwood, I. M. Eggleston and D. M. van Aalten, EMBO J., 2006, 25, 1569–1578 CrossRef CAS.
- B. Shanmugasundaram, A. W. Debowski, R. J. Dennis, G. J. Davies, D. J. Vocadlo and A. Vasella, Chem. Commun., 2006, 4372–4374 RSC.
- H. C. Dorfmueller, V. S. Borodkin, M. Schimpl, S. M. Shepherd, N. A. Shpiro and D. M. van Aalten, J. Am. Chem. Soc., 2006, 128, 16484–16485 CrossRef CAS.
- H. C. Dorfmueller, V. S. Borodkin, M. Schimpl and D. M. van Aalten, Biochem. J., 2009 Search PubMed.
- M. von Itzstein and R. Thomson, Handb. Exp. Pharmacol., 2009, 189, 111–154 Search PubMed.
- P. Miendl and H. Tuppy, Hoppe-Seyler's Z. Physiol. Chem., 1969, 350, 1088–1092.
- F. A. Shaikh and S. G. Withers, Biochem. Cell Biol., 2008, 86, 169–177 Search PubMed.
- J. M. Macdonald, R. V. Stick, D. M. G. Tilbrook and S. G. Withers, Aust. J. Chem., 2002, 55, 747–752 CAS.
- S. J. Williams, R. Hoos and S. G. Withers, J. Am. Chem. Soc., 2000, 122, 2223–2235 CrossRef.
- H. Driguez, ChemBioChem, 2001, 2, 311–318 CrossRef CAS.
- J. E. Hanson, A. P. Kaplan and P. A. Bartlett, Biochemistry, 1989, 28, 6294–6305 CrossRef CAS.
- M. A. Phillips, A. P. Kaplan, W. J. Rutter and P. A. Bartlett, Biochemistry, 1992, 31, 959–963 CrossRef CAS.
- P. A. Bartlett and C. K. Marlowe, Biochemistry, 1983, 22, 4618–4624 CrossRef CAS.
- P. A. Bartlett and M. A. Giangiordano, J. Org. Chem., 1996, 61, 3433–3438 CrossRef CAS.
- R. A. Field, A. H. Haines and E. J. T. Chrystal, Bioorg. Med. Chem. Lett., 1991, 1, 667–672 CrossRef CAS.
- V. L. Schramm, Arch. Biochem. Biophys., 2005, 433, 13–26 CrossRef CAS.
- S. G. Withers, M. Namchuk and R. Mosi, in Iminosugars as glycosidase inhibitors. Nojirimycin and beyond, ed. A. E. Stütz, Wiley-VCH, Weinheim, 1999, pp. 188–206 Search PubMed.
- R. Mosi, H. Sham, J. C. M. Uitdehaag, R. Ruiterkamp, B. W. Dijkstra and S. G. Withers, Biochemistry, 1998, 37, 17192–17198 CrossRef CAS.
- C. R. Berland, B. W. Sigurskjold, B. Stoffer, T. P. Frandsen and B. Svensson, Biochemistry, 1995, 34, 10153–10161 CrossRef CAS.
- G. E. Whitworth, M. S. Macauley, K. A. Stubbs, R. J. Dennis, E. J. Taylor, G. J. Davies, I. R. Greig and D. J. Vocadlo, J. Am. Chem. Soc., 2007, 129, 635–644 CrossRef CAS.
- J. Wicki, S. J. Williams and S. G. Withers, J. Am. Chem. Soc., 2007, 129, 4530–4531 CrossRef CAS.
- T. M. Gloster, P. Meloncelli, R. V. Stick, D. L. Zechel, A. Vasella and G. J. Davies, J. Am. Chem. Soc., 2007, 129, 2345–2354 CrossRef CAS.
- M. W. Freyer and E. A. Lewis, Methods Cell Biol., 2008, 84, 79–113 Search PubMed.
- K. Sharp, Protein Sci., 2001, 10, 661–667 CrossRef CAS.
- J. D. Dunitz, Chem. Biol., 1995, 2, 709–712 CrossRef CAS.
- A. Cornish-Bowden, J. Biosci., 2002, 27, 121–126 Search PubMed.
- A. Cooper, C. M. Johnson, J. H. Lakey and M. Nollmann, Biophys. Chem., 2001, 93, 215–230 CrossRef CAS.
- A. Cooper, Curr. Opin. Chem. Biol., 1999, 3, 557–563 CrossRef CAS.
- R. U. Lemieux, Acc. Chem. Res., 1996, 29, 373–380 CrossRef CAS.
- E. Grunwald and C. Steel, J. Am. Chem. Soc., 1995, 117, 5687–5692 CrossRef CAS.
- E. Gallicchio, M. M. Kubo and R. M. Levy, J. Am. Chem. Soc., 1998, 120, 4526–4527 CrossRef CAS.
- M. Aguilar, T. M. Gloster, I. García-Moreno, C. Ortiz Mellet, G. J. Davies, A. Llebaria, J. Casas, M. Egido-Gabás and J. M. García Fernandez, ChemBioChem, 2008, 9, 2612–2618 CrossRef CAS.
- M. Aguilar-Moncayo, T. M. Gloster, J. P. Turkenburg, M. I. García-Moreno, C. Ortiz Mellet, G. J. Davies and G. Fernádez, Org. Biomol. Chem., 2009, 7, 2738–2747 RSC.
- A. A. Edwards, J. M. Mason, K. Clinch, P. C. Tyler, G. B. Evans and V. L. Schramm, Biochemistry, 2009, 48, 5226–5238 CrossRef CAS.
- A. H. Daranas, H. Shimizu and S. W. Homans, J. Am. Chem. Soc., 2004, 126, 11870–11876 CrossRef CAS.
- A. K. Grover and R. J. Cushley, Biochim. Biophys. Acta, 1977, 482, 109–124 CAS.
- G. Hanozet, H.-P. Pircher, P. Vanni, B. Oesch and G. Semenza, J. Biol. Chem., 1981, 256, 3703–3711 CAS.
- A. Bülow, I. W. Plesner and M. Bols, Biochim. Biophys. Acta, 2001, 1545, 207–215 CAS.
- A. Lohse, T. Hardeli, A. Jensen, I. W. Plesner and M. Bols, Biochem. J., 2000, 349, 211–215 CrossRef CAS.
- J. F. Morrison, Trends Biochem. Sci., 1982, 7, 102–105 CrossRef CAS.
- J. F. Morrison and C. T. Walsh, Adv. Enzymol. Relat. Areas Mol. Biol., 1988, 61, 201–301 Search PubMed.
- W. Liu, C. J. Rogers, A. J. Fisher and M. D. Toney, Biochemistry, 2002, 41, 12320–12328 CrossRef CAS.
- D. J. Merkler, M. Brenowitz and V. L. Schramm, Biochemistry, 1990, 29, 8358–8364 CrossRef CAS.
- J. V. Schloss, Acc. Chem. Res., 1988, 21, 348–353 CrossRef CAS.
- D. W. Parkin and V. L. Schramm, Biochemistry, 1995, 34, 13961–13966 CrossRef CAS.
- M. J. Sculley, J. F. Morrison and W. W. Cleland, Biochim. Biophys. Acta, 1996, 1298, 78–86.
- V. L. Schramm, Annu. Rev. Biochem., 1998, 67, 693–720 CrossRef CAS.
- G. Legler, in Iminosugars as glycosidase inhibitors. Nojirimycin and beyond, ed. A. E. Stütz, Wiley-VCH, Weinheim, 1999, pp. 31–67 Search PubMed.
- M. P. Dale, H. E. Ensley, K. Kern, K. A. R. Sastry and L. D. Byers, Biochemistry, 1985, 24, 3530–3539 CrossRef CAS.
- G. Legler and M.-T. Finken, Carbohydr. Res., 1996, 292, 103–115 CrossRef CAS.
- M. T. H. Axamawaty, G. W. J. Fleet, K. A. Hannah, S. K. Namgoong and M. L. Sinnott, Biochem. J., 1990, 266, 245–249 CAS.
- L. P. McIntosh, G. Hand, P. E. Johnson, M. D. Joshi, M. Körner, L. A. Plesniak, L. Ziser, W. W. Wakarchuk and S. G. Withers, Biochemistry, 1996, 35, 9958–9966 CrossRef CAS.
- T. M. Gloster, S. J. Williams, S. Roberts, C. A. Tarling, J. Wicki, S. G. Withers and G. J. Davies, Chem. Commun., 2004, 1794–1795 RSC.
- J. R. Knowles, Crit. Rev. Biochem. Mol. Biol., 1976, 4, 165–173 Search PubMed.
- K. Brocklehurst, Protein Eng., Des. Sel., 1994, 7, 291–299 CrossRef CAS.
- S. K. Konstantinidis, M. L. Sinnott and B. G. Hall, Biochem. J., 1993, 291, 15–17.
- J. T. Bolin, D. J. Filman, D. A. Matthews, R. C. Hamlin and J. Kraut, J. Biol. Chem., 1982, 257, 13650–13662 CAS.
- F. Payan, P. Leone, S. Porciero, C. Furniss, T. Tahir, G. Williamson, A. Durand, P. Manzanares, H. J. Gilbert, N. Juge and A. Roussel, J. Biol. Chem., 2004, 279, 36029–36037 CrossRef CAS.
- S. Sansen, C. J. De Ranter, K. Gebruers, K. Brij, C. M. Courtin, J. A. Delcour and A. Rabijns, J. Biol. Chem., 2004, 279, 36022–36028 CrossRef CAS.
- D. R. Houston, K. Shiomi, N. Arai, S. Omura, M. G. Peter, A. Turberg, B. Synstad, V. G. H. Eijsink and D. M. F. van Aalten, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 9127–9132 CrossRef CAS.
- Y.-K. Li and L. D. Byers, Biochim. Biophys. Acta, 1989, 999, 227–232 CAS.
| This journal is © The Royal Society of Chemistry 2010 |
