Temporal and mechanistic tracking of cellular uptake dynamics with novel surface fluorophore-bound nanodiamonds
Amanda M.
Schrand
*a,
Jonathan B.
Lin
a,
Suzanne Ciftan
Hens
b and
Saber M.
Hussain
*a
aApplied Biotechnology Branch, Human Effectiveness Directorate, AFRL/RHPB, 2729 R Street, Bldg 837, Wright-Patterson AFB, OH 45433-5707, USA. E-mail: amanda.schrand@wpafb.af.mil; saber.hussain@wpafb.af.mil; Fax: +937-904-9610; Tel: +937-904-9514 Tel: +937-904-9517
bInternational Technology Center, Raleigh, NC 27617, USA Web: www.itc-inc.org
First published on 27th September 2010
Abstract
Nanoparticles (NPs) offer promise for a multitude of biological applications including cellular probes at the bio-interface for targeted delivery of anticancer substances, Raman and fluorescent-based imaging and directed cell growth. Nanodiamonds (NDs), in particular, have several advantages compared to other carbon-based nanomaterials – including a rich surface chemistry useful for chemical conjugation, high biocompatibility with little reactive oxygen species (ROS) generation, physical and chemical stability that affords sterilization, high surface area to volume ratio, transparency and a high index of refraction. The visualization of ND internalization into cells is possible via photoluminescence, which is produced by direct dye conjugation or high energy irradiation that creates nitrogen vacancy centers. Here, we explore the kinetics and mechanisms involved in the intracellular uptake and localization of novel, highly-stable, fluorophore-conjugated NDs. Examination in a neuronal cell line (N2A) shows ND localization to early endosomes and lysosomes with eventual release into the cytoplasm. The addition of endocytosis and exocytosis inhibitors allows for diminished uptake and increased accumulation, respectively, which further corroborates cellular behavior in response to NDs. Ultimately, the ability of the NDs to travel throughout cellular compartments of varying pH without degradation of the surface-conjugated fluorophore or alteration of cell viability over extended periods of time is promising for their use in biomedical applications as stable, biocompatible, fluorescent probes.
1. Introduction
A multitude of nanoparticles (NPs) are being examined for their suitability to interact with biological entities at the nano-surface level in areas such as drug delivery, detoxification, sensing, and imaging.1–3 As the number of available NPs increases, it becomes a priority to understand the mechanisms through which they may enter the intracellular space. The majority of studies on the cellular uptake of NPs have cited endocytosis as the mechanism.4–8 For example, carbon nanotubes (CNTs) complexed to proteins or DNA were shown to be uptaken viaclathrin-dependent endocytosis.4–6 In other studies, quantum dot-multi-walled CNT conjugates7 and multi-photon carbon dots8 were internalized by endocytosis after 1 h at 37 °C compared to little or no uptake at 4 °C. In contrast, non-endocytic or non-phagocytic uptake mechanisms based on diffusion, adhesion, or other interactions may play a role in internalization. For example, the uptake of fluorescent fluorescein isothiocyanate (FITC)-conjugated CNTs into the cytoplasm and nucleus of cells appeared to be energy-independent,9,10 while fluorescent polystyrene microspheres did not localize to membrane-bound structures within pulmonary macrophages or red blood cells.11 Although the exact mechanism of NP entry is difficult to pinpoint, other studies show that CNTs localized to intracellular vesicles in human keratinocytes12 or fullerenes to mitochondria in COS-7 monkey kidney cells after 24 h.13 More recently, Zhang and Monteiro found that quantum dots were internalized into early endosomes and then transferred to late endosomes or lysosomes with the endocytic pathway primarily regulated by the G-protein-coupled receptor/scavenger receptor in human epidermal keratinocytes.14 In this regard, it is essential to track the location of NPs inside cells over time to evaluate accumulation kinetics. Factors found to influence NP uptake by cells include size, surface charge, concentration, time, temperature, and cell line.7,8,15–17Detonation NDs are very attractive candidates for intracellular studies due to their ability to be produced with high purity, low cost, high yield synthesis methods,18–20 sterilized by either filtration or autoclave methods, ease of surface functionalization, lack of reactive oxygen species (ROS) generation, pH stability, stable fluorescence and their greater inherent biocompatibility with cells in culture17,18,21–23 compared to other nanomaterials such as carbon nanotubes or quantum dots.24 Consequently, NDs have been used for drug-delivery applications and as enterosorbents.25 Further, in vivo biodistribution26 and anticancer studies27 show promising preliminary results.
As imaging probes, NDs can be viewed with multiple methods of optical and spectral detection and associated imaging techniques (e.g., N-vacancy induced fluorescence, optical transparency in the visible wavelength range, high index of refraction, and Raman scattering signal).18,20,28,29,31–34,69 The further surface functionalization of NDs through attachment of biomolecules has been demonstrated by direct physisorption.35,36 This can be accomplished either by indirectly linking amines to physisorbed polylysine37 or by grafting silane as a linker to couple amino acids or bioconjugates to the ND surface.38,39 Covalent coupling of the dye molecule, TAMRA – through the carbon surface of NDs, using standard wet chemical synthesis methods – allowed for dual visualization of NDs and streptavidin.40,41
In this study, the rhodamine-conjugated NDs (ND-T), as mentioned above, were used to characterize mechanistic uptake dynamics in neuroblastoma cells. This neuronal cell phenotype was chosen to simulate both the deliberate targeting of NPs to the nervous system or accidental exposure and subsequent translocation from sites of administration.42 NDs labeled with pH-independent TAMRA dye showed photostability and high brightness, likely due to the NDs' high index of refraction. ND-T is positively charged and has a high degree of colloidal stability in aqueous solution.40 Imaging with transmission electron microscopy (TEM) was used to examine uptake and localization over time, and the uptake pathway was corroborated by using chemical inhibitors. Further staining of subcellular organelles and imaging with confocal microscopy was utilized to temporally track the uptake and localization of ND-T. Through these studies, a better understanding of NP uptake is gleaned towards further surface-tailoring of intelligently designed nanostructures.43,44
2. Materials and methods
Fluorescent ND synthesis and characterization
Detonation NDs (NDs, >98% purity, designated I6, supplied by New Technologies, Co., Chelyabinsk, Russian Federation) were functionalized by ITC as described in40 by amination of the ND and coupling to the reactive NHS functionalized TAMRA to produce the singly conjugated particle TAMRA-ND (ND-T). X-ray photoelectron spectroscopy (XPS) was performed using a Kratos Axis Ultra with a monochromatic Al kα X-ray source and a silicon wafer as a supporting substrate. The survey and high resolution scans were performed at 160 eV pass energy with 1 eV step and 10 eV pass energy with 0.1 eV step, respectively. The peak position calibration was done against Si 2p3/2 @ 99.3 eV.45 The collected data were processed with CasaXPS software. A Shimadzu FTIR-8300 spectrometer was used for FTIR analysis with a fixed quantity of ND sample mixed with KBr and compressed at up to 150 kg/cm2 into plates 0.5–0.7 mm thick. Tablets were placed in an IR vacuum cell and heated at 100 °C under vacuum (1 × 10−2 torr) for 2 h inside the IR cuvette in order to remove traces of water. After this procedure, FTIR spectra were recorded without exposure of the samples to air in order to avoid any influence due to the atmospheric water.The amount of TAMRA in the ND-T samples obtained by the chemical reaction between aminated ND and NHS-TAMRA was measured by comparison of the nonreactive form of TAMRA. The experimental procedure performed to quantify the binding of physisorbed and chemically bound TAMRA were previously reported;40 however, in this paper we show the binding curves. An amount of 100 μL of I6-NH2 (10 mg/mL in 0.1 M bicarbonate buffer pH 8.1) was mixed with 200 μL of 0.1M bicarbonate buffer (pH 8.1). Stock solutions of 16.8 mM TAMRA and 9.89 mM TAMRA-NHS were prepared in DMSO as measured by UV-Vis at the peak absorbance of 551 nm. Each series consisted of four reactions using 5, 15, 20, and 40 μL of TAMRA or 2, 10, 20, and 40 μL of TAMRA-NHS added to each ND solution. The solutions were kept at room temperature under mixing for 90 min. Next, the ND was pelleted using 6,000 rpm for 5 min and the supernatant was collected. The ND pellet was then resuspended in 0.1 M bicarbonate buffer (pH 8.1) and centrifuged again with removal of the supernatent. The amount of dye removed from the ND was measured by UV-Vis and the total amount of TAMRA removed was calculated.
ND-T was prepared as a stock solution at a concentration of 7.5 mg/mL in water, which was then further diluted in sterile water to 1 mg/mL before use in cell culture experiments. Verification of the size and morphology was performed with fluorescent microscopy (Olympus IX71 Inverted Research Microscope) and transmission electron microscopy (TEM, Hitachi H-7600, 110 kV).
Cell culture
Neuroblastoma (N2A) cells, a neuronal phenotype, were purchased from ATCC (Neuro-2a line, CCL-131). Cells were grown in an atmosphere of 5% CO2 and 37 °C according to standard cell culture techniques.17,19,46–48Growth media for the neuroblastoma cells was DMEM/F12 supplemented with 10% normal fetal bovine serum (FBS). Growth media for the human keratinocyte (HaCaT, ATCC) cells was RPMI-1640 supplemented with 10% FBS, and growth media for the rat alveolar macrophages (ATCC, NR8383 line, CRL-2192) was Ham's Nutrient Mixture F-12 K (Kaughn's Modification) media (ATCC) supplemented with 20% FBS. All cell culture media also contained 1% Pencillin-streptomyocin (ATCC). Other cell culture supplies included 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT, Sigma Chemical Company, St. Louis, MO), 10x Phosphate buffered saline (pH 7.4), and 2.5% trypsin (Gibco Invitrogen™ Corporation, Carlsbad, CA).Biocompatibility assay
For biocompatibility evaluation, all cells were seeded in 6-well plates at a concentration of 5 × 105cells/mL. The 3-[4,5-Dimethyl-thylthiazol-2-yl]-2,5-diphenyl-tetrazolium bromide (MTT) assay was conducted to assess cellular viability based on mitochondrial function. After 30 min of incubation with MTT, a purple color developed within the cells, indicating the cleavage of the tetrazolium salt (MTT) by active mitochondria in live cells. The purple-colored product (formazan crystals) was extracted into solution with acidified isopropanol for homogeneous staining, and the absorbance was measured on a Spectromax 190 microplate reader from 570–630 nm after centrifugation to remove the NPs. The percent reduction of MTT was compared to controls (cells not exposed to NDs), which represented 100% MTT reduction.Transmission electron microscopy of cellular uptake
N2A cells were processed for TEM according to Schrand et al.48 Briefly, the cells were seeded in 100 mm tissue culture dishes, and then dosed with NDs at 85% confluency. Twenty-four hours later, the cells were fixed in 2.5% paraformaldehyde/gluteraldehyde in PBS for 2 h. Thereafter, the cells were post-fixed with 1% osmium tetroxide for 1 h, and then the cells were scraped from the plate and centrifuged. The cells were dehydrated using increasing concentrations of ethanol with three changes of 100% ethanol. The samples were then placed in 100% LR White resin and cured overnight at 60 °C in BEEM®capsules. The samples were thin-sectioned on a Leica ultramicrotome at a thickness of 50–100 nm, collected on formvar/carbon-coated TEM grids and imaged using a Hitachi H-7600 TEM at 110kV.Morphological evaluation with light microscopy
For morphological evaluation, N2A cells were grown to 80% confluency in two-chambered glass slides for phase contrast light microscopy (Olympus IX71 inverted light microscope with QCapture imaging software). Subsequently, cell cultures were dosed with freshly prepared ND-T at concentrations from 0–100 μg/mL in cell culture media without serum for N2A or HaCat cells, or media with 10% serum for macrophages to reduce proliferation.Confocal localization and uptake studies
Confocal imaging was performed on a BD Pathway 435 instrument with associated AttoVision software. In live cell fluorescent confocal experiments, Hoechst 33342 nuclear dye (Biomol, Plymouth Meeting, PA) was used as a counterstain. The early endosome stain EEA-1 (Abcam, Cambridge, MA) was utilized at a working concentration of 1–4μg/mL in exposure media. After 1 h of incubation, the cells were washed 3× with warmed exposure media. LysoTracker Green lysosome stain (Invitrogen/Molecular Probes, Eugene, OR) was probed under the FITC filter for co-localization experiments according to the manufacturer's protocol.Image Data Explorer software was utilized to quantify the uptake of fluorescent NDs. A compound macro was set up to collect images for an entire 96 well plate. Afterwards, the analysis function of the Confocal microscope segmented the image based on differences in fluorescence, distinguishing areas of Rhodamine and Hoechst intensity from background fluorescence. Finally, the Image Data Explorer quantified these areas, providing a numerical value of relative fluorescent units. This value represents the quantity of ND-Ts present and is proportional to cellular ND-T uptake since all wells were seeded at similar densities.
Chemical inhibitors
For modulating cellular uptake, known chemical inhibitors of endocytosis (Dynasore) and exocytosis (Brefeldin A, BFA) were utilized. Dynasore is a known inhibitor of dynamin, a protein required for clathrin-dependent vesicle formation,49,50 while Brefeldin A is a known lactone antibiotic that interferes with the merging of endosomes and lysosomes, thereby resulting in an accumulation of proteins and extracellular transports within the cell.51,52 These chemicals were co-incubated with the NPs at low doses of 20 μM and 3 μg/mL respectively to interfere with normal processes of uptake and migration.Statistical analysis
The above biochemical assays were performed in triplicate and the results were presented as mean ± standard deviation in comparison to control values. The data were subjected to statistical analysis by one-way analysis of variance (ANOVA) followed by Tukey-Kramer's procedure for multiple comparisons (pHStat Excel add-in). A value of p < 0.05 was considered significant and marked with asterisks (*) on graphs.3. Results and discussion
Fluorescent tetramethylrhodamine (TAMRA)-labeled NDs (ND-T) were previously characterized as a stable aqueous colloidal suspension with a zeta potential of +40 mV, a primary size of ∼ 4 nm, and after suspension in water, an average aggregate size of ∼150 nm.40 Since NDs are a good sorbent for biomolecules, the relative amounts of chemically bound and physisorbed TAMRA on the ND surface were measured. Most experiments using ND as a fluorescent tracer require chemical bonding of the dye to the ND surface since this complex is stronger and more stable than physisorption. The chemical reaction of the amine functionality of I6-NH2 with the NHS ester group of TAMRA-NHS produces a chemically stable amide bond. In order to show that the conjugation reaction is actually taking place due to the amine functionality on I6-NH2 with the amine reactive NHS ester of TAMRA, the reaction was completed along with the control reaction using TAMRA without the NHS reactive group over a series of concentrations of dye. Using the same amount of I6-NH2 along with increasing dye concentration, the amount of dye removed from the ND through repeated centrifugation pelleting and washing steps was measured. In this case, the majority of the dye was removed after two washes.The corresponding Langmuir isotherms are shown in Fig. 1. It can be seen that I6-NH2 appears to chemically bind TAMRA (green line) using the TAMRA-NHS reactive moiety, since TAMRA dye used without the reactive ester function has fewer moles of dye bound to the ND. In addition, we can see that TAMRA physisorption (blue line) begins to saturate at a higher dye concentration. Furthermore, as previously reported, the ND-T sample using NHS-TAMRA appeared red, whereas the ND mixed with unreactive TAMRA had no apparent coloration.40
 | ||
| Fig. 1 Plot of the amount TAMRA bound to I6-NH2 as a function of amount of TAMRA removed or in equilibrium for the reaction (green) and the control (blue). Measurements were an average of three data sets. The deviation in each measurement of bound TAMRA is shown with error bars. | ||
ND-T product was characterized using XPS and FTIR after extensive removal of unbound TAMRA dye from the ND surface. XPS is reported (Fig. 2) for the starting material (I6), the amine functionalized ND (ND-NH2), and the dye conjugate (ND-T). The peaks at 399.8 and 399.7 in all samples are indicative of nitrogen in a sp3 type environment (amine).53 The amination reaction resulted in a dramatic increase in the overall content of sp3 type nitrogen, which is due to the formation of amine species on the surface of the ND. The ND-T product had an amine peak at 399.7, which may be attributed to the amide bond between ND and TAMRA, as well as the two nitrogen sp3groups from TAMRA. The peak at 400.8 eV in the first two samples is indicative of nitrogen in a sp2 type environment (imine).54 The peak at 403.6 eV is indicative of N–O species (nitro, nitroso).55,56 Additional peaks were seen in the ND-T product which can be attributed to the conjugated ring system of TAMRA and the charged N in TAMRA.
 | ||
| Fig. 2 XPS spectra of (a) ND-I6, (b), ND-NH2 and (c) ND-T demonstrating progression of surface functionalization (amine, imine, nitro, nitroso and amide bond formation). | ||
The FTIR spectra are compared for the samples taken in air (Fig. 3). By comparing the ND-NH2 and ND-T spectra, a difference in the intensity distribution of peaks is seen, shown as red arrows. The starting ND I6 sample and ND-T have similarities in the stretching modes of C–H (ν 2800–2900 cm−1) and C–H bending mode (1460 cm−1). The similarity is caused by the addition of the organic moeity TAMRA. Bonded water may be seen in the region between 3200 and 3500 cm−1 and at 1635 cm−1. All three species have oxygen-containing groups, such as: ≥C–OH (3200–3600 cm−1 - ν O–H in water, hydroxyl groups in carboxylic or tertiary alcohol). Whereas I6 has a significant peaks for ≥C–O–C≤ (ν 1100–1370 cm−1 - in ether, acid anhydride, lactones, epoxy groups), these groups were removed in the reduction reaction. The >C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/b_char_e001.gif) Ogroup (1700–1865 cm−1 in ketonic, carboxylic, acid anhydrides groups, ester and lactones) was also reduced and is not present in the aminated ND material. For ND-NH2 the O–H and N–H bands (ν1635 cm−1) are indicative of an amine group in ND-NH2. For ND-T, this region is larger relative to the C–H and O–H frequencies at 1384 cm−1. This increase in hydrocarbons can be caused by TAMRA. Also, for ND-T this band is possibly broadened due to the amide bond between TAMRA and ND-NH2 (shoulder at 1550 cm−1). Other peaks for the amide group include 3320 and 3360 cm−1 for the amide N–H bond, which increased relative to ND-NH2.
Ogroup (1700–1865 cm−1 in ketonic, carboxylic, acid anhydrides groups, ester and lactones) was also reduced and is not present in the aminated ND material. For ND-NH2 the O–H and N–H bands (ν1635 cm−1) are indicative of an amine group in ND-NH2. For ND-T, this region is larger relative to the C–H and O–H frequencies at 1384 cm−1. This increase in hydrocarbons can be caused by TAMRA. Also, for ND-T this band is possibly broadened due to the amide bond between TAMRA and ND-NH2 (shoulder at 1550 cm−1). Other peaks for the amide group include 3320 and 3360 cm−1 for the amide N–H bond, which increased relative to ND-NH2.
 | ||
| Fig. 3 FTIR spectra of I6 (Green), ND-NH2 (Blue), ND-T (Red) taken in air. Bands are shown at their corresponding wavenumber and bonding modes. Red arrows indicate an increase in intensity of ND-T with respect to ND-NH2. | ||
Ultimately, the chemical characterization results demonstrate that the final product ND-T is attached to the ND surface through a strong chemical bond. Representative fluorescent (Fig. 4A) and transmission electron micrographs (TEM, Fig. 4B) of a 1 mg/mL ND-T stock solution are shown in Fig. 4. Although the primary size of the ND-T was ∼2–10 nm (Fig. 4B), small aggregates could be found after introduction into cell culture media (Fig. 4B). The issue of dispersion to aggregate sizes less than 50 nm in various physiological solutions has been discussed elsewhere.57–61
 | ||
| Fig. 4 Images of stock solution of 1 mg/mL fluorescent NDs (ND-T). (A) Fluorescent image, (B) Transmission electron micrograph demonstrating fluorescence and primary ND size/morphology, respectively. | ||
To determine the biocompatibility of ND-T in both rodent and human cell lines, changes in cell viability after 24 h of incubation with 5–100 μg/mL ND-T were assessed with the MTT assay (Fig. 5). Statistical analysis indicates that N2A, HaCat, and macrophage cell viabilities do not significantly differ from the control at any of the concentrations tested. Additionally, morphological examination did not show any alterations in cell shape compared to the controls (Fig. 5). Together, these biochemical and visual findings support the low toxicity of ND-T after 24 h even at increasing doses with concurrent cellular accumulation.
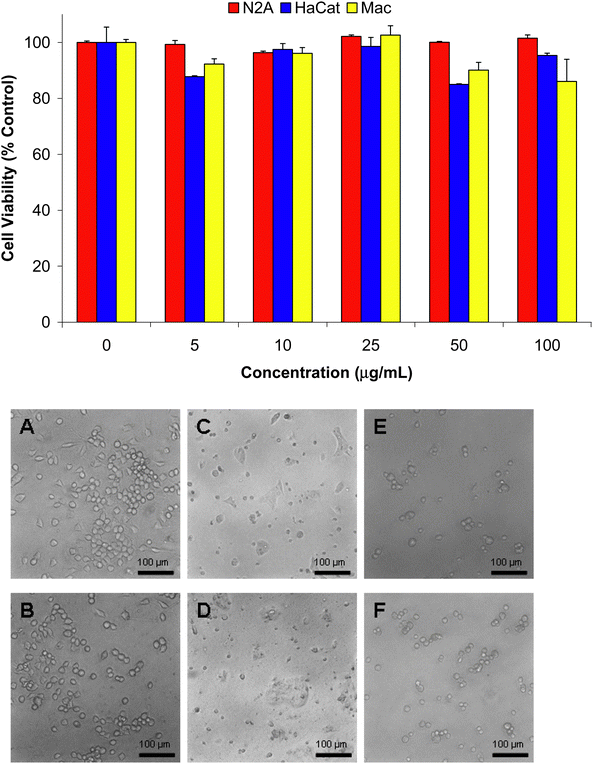 | ||
| Fig. 5 MTT viability assessement and morphological evaluation of N2A, HaCat, and macrophage cells after 24 h of incubation with 0–100 μg/mL of ND-T. Triplicate experiments show no significant difference in viability after exposure to ND-T compared to untreated controls. Phase contrast light microscopy of (A,C,E) Controls and (B,D,F) Cells incubated with 100 μg/mL ND-T. (A,B) N2A cells, (C,D) HaCat cells, and (E,F) Macrophages. There were no morphological changes compared to the controls. | ||
Transmission electron microscopy (TEM) was performed on N2A cells in order to initially investigate the time-dependent uptake of ND-T into cells as well as their localization. The TEM studies enabled both verification of uptake and higher resolution sub-cellular localization of ND-Ts. Images in Fig. 6 show ND-T outside cells (white arrows), internalized into membrane-bound structures resembling early endosomes inside cells (black arrows) as well as free in the cytoplasm after lysosomal processing (Fig. 6I). The presence of ND-T aggregates increases over time from 1h to 6 h (Fig. 6A–H) compared to 24 h where larger pockets were found (Fig. 6I–L). The ND aggregates inside the intracellular vacuoles were ∼500 nm in size. Several images suggest that the NDs, after coming into contact with the cell membrane, are brought inside by an endocytic mechanism where they remain close to the edge of the membrane, but do not appear to enter the nucleus. Additionally, the degradation of the membrane-bound structure containing the NDs was found in some areas after 24 h (Fig. 6I). Confirmation of the increased uptake of ND-T over time was demonstrated with confocal microscopy after static incubation with 10 μg/mL of ND-T over 1–24 h (Fig. 7A–C).
 | ||
| Fig. 6 Internalization and localization of NDs into N2A cells with TEM. (A–D) After 1h incubation, (E–H) After 6 h incubation, (I–L) After 24 h incubation. Notice the time-dependent accumulation of ND-T both in the cytoplasm and intracellular vacuoles. | ||
 | ||
| Fig. 7 Representative confocal microscope images of N2A cells incubated with ND-T (red) and counterstained with and Hoechst dye (blue) for nuclear staining. Time-dependent increases in ND-T accumulation at a static dose of 10 μg/mL from 1–24 h. (A) 1h incubation, (B) 3 h incubation and (C) 24 h incubation with ND-T. Images were taken at 60× magnification. | ||
To further investigate ND-T intracellular trafficking based on specific uptake and release mechanisms, chemical inhibitors of endocytosis and exocytosis were employed in conjunction with confocal microscopy. The chemical compounds Dynasore and Brefeldin A (BFA) were chosen to regulate entry or exit of the NDs, respectively. Dynasore inhibits dynamin, a protein required for clathrin-dependent vesicle formation.49,50BFA interferes with the merging of endosomes and lysosomes, thereby resulting in an accumulation of proteins and extracellular transports near organelles such as the rough endoplasmic reticulum.51,52 When ND-T uptake was chemically inhibited with Dynasore (Fig. 8B) or retention induced with BFA (Fig. 8C), there were visible and quantifiable reductions or increases in fluorescent intensity, respectively, relative to untreated cells (Fig. 8A).
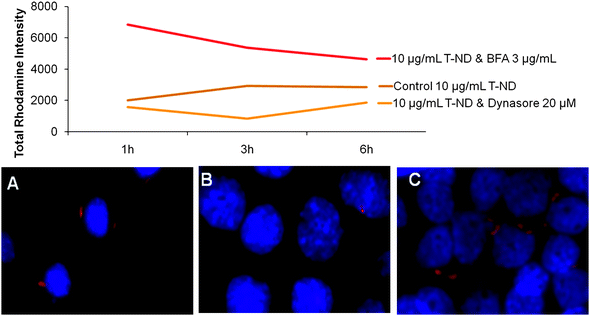 | ||
| Fig. 8 Effect of endocytosis and exocytosis inhibition on the uptake and release dynamics of ND-T in N2A cells. Cells were dosed with 10 μg/mL ND-T for 3 h with (B, C) or without (A) direct incubation with chemical inhibitors and assessed with confocal microscopy. Relative fluorescent intensities were higher when exocytosis was blocked with the chemical inhibitor BFA (red line) compared to the control (brown line) and reduced fluorescent intensities were obtained after blocking endocytosis with the chemical inhibitor Dynasore (orange line). Representative images show visual depiction of control ND-T uptake (A) compared to reduced ND-T uptake after treatment with endocytosis inhibitor Dynasore (B) and increased ND-T uptake after blocking exocytosis with chemical inhibitor BFA (C). | ||
Because an endocytic mechanism appeared to be at work, the co-localization of ND-T in distinct subcellular locations was probed with organelle-specific stains for early endosomes and lysosomes in a chronological study. Early endosomes are observed during the early stages of clathrin-dependent endocytosis as membrane vesicles pinch off containing extracellular transports including NPs. Lysosomes are single-membrane compartments found in most eukaryotic cells that are responsible for the breakdown of materials.62 In the case of endocytosis, lysosomes develop gradually from late endosomes, which are vesicles that initially carry materials into the cell. The coalescence of the endosome with the low pH environment of the lysosome causes the degradation and release of the contents. Therefore, we expect to observe early localization of ND-T to endosomes, followed by localization to lysosomes, and finally to the cytosol.
In cells fixed and stained for detecting early endosomes, control N2A cells show diffuse green staining with some punctate spots that represent invaginating early endosomes (Fig. 9A). After 1 h, green punctate dots can still be observed; however, there is some overlap of red signal from ND-T with the green dots, suggesting that there is early and rapid localization to early endosomes (Fig. 9B). After 3 h, there is increased ND-T density within the early endosomes (Fig not shown).
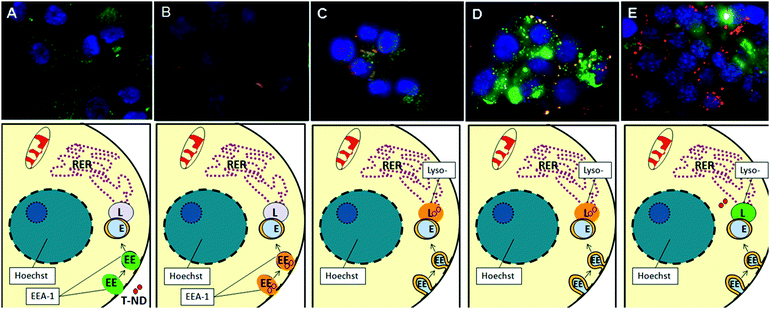 | ||
| Fig. 9 Confocal microscopy of N2A cells for co-localization of ND-T (10 μg/mL) and early endosomes or lysosomes after various time points. Labels: ND-T (red), Hoechst nuclear dye (blue) and LysoTracker/EEA1 (green). Combined red and green signals, indicative of co-localization of ND-T in lysosomes or early endosomes, appear in yellow or orange. (A) Control with EEA1, (B) 1 h, stained for EEA1, (C) 3 h, stained for LysoTracker (D) 6 h, stained for LysoTracker, (E) 24 h, stained for LysoTracker. Notice that not all ND-T is localized to lysosomes, but some NDs are free in the cytoplasm and not all lysosomes contain ND-T. Images were taken at 60× magnification. EEA1 at 3 h, 6 h, 24 h and LysoTracker at 0 h, 1 h were omitted for clarity and ease of presentation. | ||
In cells fixed and stained for lysosomes, control N2A cells show diffuse green staining with some very small punctate spots indicative of the lysosomes (Fig not shown). However, there was very little localization to lysosomes after a short incubation with 10 μg/mL of ND-T for 1 h (Fig not shown). After 3 h, there was some overlapping of the red signal from ND-T and the green signal representative of lysosomes resulting in yellow spots, suggesting that ND-T co-localized with the lysosomes (Fig. 9C). After 6 h, the density of this overlap greatly increases, suggesting that movement to the lysosomes follows early endosomes localization (Fig. 9D). After 24 h, most of the ND-T appeared to be released from the lysosomes and was found in the cytoplasm as red aggregates while the lysosomes remained stained in green (Fig. 9E).
4. Discussion and conclusions
In summary, we have demonstrated the time-dependent uptake and localization of ND-T with significant accumulation while maintaining high biocompatibility (Table 1). These NDs are able to maintain low toxicity in vitro while maintaining strong surface stability of its protein conjugates and bright fluorescence, making them strong candidates for applications in nano-biotechnology and nano-biomedicine. Based on our TEM and Confocal imaging data, we propose that the uptake mechanism is endocytic as shown schematically in Fig. 9.

|
Similar to our previous results with raw, acid-, or base-purified detonation NDs,19 this study with novel ND-Ts found internalization by both animal and human cell lines. Moreover, cells with internalized NDs were able to maintain high viabilities at concentrations up to 100 μg/mL for 24 h even with NDs from different vendors.17,19 The characteristics of this novel ND-T allowed dual visualization with both fluorescent confocal microscopy and transmission electron microscopy thereby providing a strong particle tracking method inside of cells.
While other groups have utilized similar techniques to track the final ND destination in cells,29,63 the use of chemically stable, bright dye-conjugated NDs allows for versatile tailoring of dyes onto the NP surface without causing chemical or structural changes in the NP itself. Dye conjugated NDs are easily synthesized using standard conjugation reactions and provide a complementary method compared to internally photoluminescent NDs. While innately photoluminescent NDs, created by high energy methods do not photoblink or photobleach, they currently have a narrow range of available photoluminescent colors and their brightness cannot be tuned. Furthermore, their photoluminescence may be particle-size dependent.
Localization
In this study, we tracked ND-T throughout the cytoplasm of cells and revealed time-based localization to intracellular vacuoles ∼500 nm in size, which include early endosomes, lysosomes, and generalized aggregation in the cytoplasm between 1–24 h. The time-based and sequential progression of ND-T through cellular compartments strongly implicates clathrin-mediated endocyotis as a major uptake pathway.Mechanism
After coming into contact with the outside of the cell membrane, ND-T appeared to be taken inside the cell by an endocytic mechanism, remaining close to the edge of the membrane in early endosomes. Other recent studies have shown the uptake of carbon nanomaterials by both endocytosis4–8 and energy-independent and non-endocytic pathways9,10 into the cytoplasm and nucleus of cells. In order to corroborate the clathrin-dependent endocytosis pathway, Dynasore was utilized to inhibit this process. Our confocal imaging data indicates that inhibition of this pathway seemed to reduce quantified uptake, as measured by Rhodamine intensity. As such, this data provides further evidence to suggest that ND-T rely at least in part on the clathrin-mediated endocytosis pathway for their uptake.Routing to lysosomes/localization to cytoplasm
Although ND-T was routed for degradation by lysosomes, the process was not successful. After 24 h, most of the ND-T remained in the cytoplasm (Fig. 9E), which suggests that the ND-T may either be released from lysosomes by this time or newly internalized NDs may have entered the cells without yet localizing to lysosomes, potentially by non-endocytic mechanisms. The strong intracellular localization of the ND-T throughout the cytoplasm of the cells (Fig. 9E) corresponded well with previous studies of non-fluorescent or electron-beam irradiated diamond NPs that localized to the cytoplasm, but were restricted from the nucleus.18,19,32 The use of Brefeldin A confirms the interaction between early endosomes and lysosomes during the early ND uptake pathway. Cells treated with this chemical inhibitor exhibited increased accumulation, which seems to be caused by the prevention of merging of early endosomes with lysosomes in the natural degradation process. This may result in a delay in the internal processing at the endosome stage, which explains the increased accumulation observed in the Confocal imaging studies.The size of the ND clusters may regulate the uptake mechanism and intracellular fate with smaller NDs localizing to the cytoplasm.63 However, the general ability of the NDs to deagglomerate in the cytoplasm provides a viable mechanism for intracellular release, which is particularly relevant for the intentional design of sustained endosomal release. In this case, predestination of the surface-functionalized NDs to the cytoplasm over time may increase the efficiency of targeted biomolecules in gene and drug delivery and was also explored by Faklaris et al.63
Long-term imaging/static uptake and accumulation
TEM images support the notion of continued uptake beyond 24 h because the plasma membrane shows invaginations and many pockets of ND-Ts, which were located at the edge of the plasma membrane (Fig. 6I–L). The degradation of the membrane-bound structure containing the NDs was found in some areas with TEM imaging after 24 h, which may explain why there is a reduction in the co-localization of fluorescent signals for the ND-Ts and the lysosome stain at 24 h (Fig. 6I, red arrow). Since photobleaching of ND-T was ruled out, another explanation for the change in fluorescent signals could be the continued settling, contact, and internalization with the cells over time. This could have been due to static dosing conditions, because at 24 h, the plasma membrane still appears to be invaginating to internalize more NDs (Fig. 6I–L). Although innately photoluminescent NDs have been used in continuous cell imaging, it is also possible for ND-T to be used as a cellular tracking label in static images beyond a 24 h time period without TAMRA photobleaching nor destabilization of the ND surface conjugation. This may prove useful in exploration of long-term ND-T cellular dynamics.Several recent studies have demonstrated the use of NDs as carriers for DNA,64,65proteins30,66–69 and drugs.28,36 However, without stable covalent bonds, many surface-conjugated NPs can lose their desirable therapeutic properties if these weak bonds are broken during the delivery process. As a result, NPs with covalently-bonded surface attachments may require a mechanism for unloading of drugs. Nonetheless, controlling NP uptake and specific targeting of NPs to subcellular locations is not yet optimized due to (1) containment within endosomes and (2) creation of temporary holes in the plasma membrane leading to toxicity.
In this study, the high photostability of the ND-T complex permitted its continuous visualization during entrapment in endosomes and then sequestering to lysosomes. TAMRA is a pH independent fluorophore, and this property appears to be retained in the ND-T complex that allowed the continued visualization of ND in the lysosome's low pH environment and preserved visualization upon release into the cytoplasm. To facilitate accurate targeting of NPs to specific locations, novel combinations of materials and surface conjugation to antibodies or the transactivating regulatory protein Tat are being explored.70–73 However, advances in producing multifunctional NPs and comparisons between in vitro and in vivo testing for biocompatibility74 are required before the full potential of NPs can be realized in biomedical applications. Furthermore, while surface functionalization may ensure delivery of a particular conjugated protein to the viscera, there are still many technical challenges for the development of advanced NP applications, such as increasing dispersion in physiological solutions, reducing endogenous fluorescent scattering of cells or autofluorescence of tissues, ensuring the functionality of attached biomolecules, and overcoming the small size of nuclear pores. Nonetheless, as these technical challenges are resolved, we may begin to exploit nanodiamonds' immense applications in the fields of nano-biotechnology and nano-biomedicine as highly stable and biocompatible, fluorescent probes.
Acknowledgements
The authors would like to thank Dr Olga Shenderova and Dr Garry Cunningham of ITC, V. Kuznetsov for helpful discussions, Sergey Moseenkov for taking FTIR spectra and Dr Laura Braydich-Stolle of AFRL/RHPB. A.M.S. received funding from the National Research Council (NRC) Fellowship program funded by the Joint Science and Technology Office for Chemical and Biological Defense (JSTO-CBD), a program administered by the Defense Threat Reduction Agency (DTRA). S.H. is thankful for the support from the Army Research Office under grant W911NF-04-2-0023.References
- R. Martın, M. Alvaro, J. R. Herance and H. Garcia, Fenton-Treated Functionalized Diamond Nanoparticles as Gene Delivery System, ACS Nano, 2010, 4, 65–74 CrossRef.
- L. Zhang, J. M. Chan, F. X. Gu, J.-W. Rhee, A. Z. Wang, A. F. Radovic-Moreno, F. Alexis, R. S. Langer and O. C. Farokhzad, Self-Assembled Lipid Polymer Hybrid NPs: A Robust Drug Delivery Platform, ACS Nano, 2008, 2, 1696–1702 CrossRef CAS.
- M. Chen, E. D. Pierstorff, R. Lam, S.-Y. Li, H. Huang, E. Osawa and D. Ho, Nanodiamond-Mediated Delivery of Water-Insoluble Therapeutics, ACS Nano, 2009, 3, 2016–2022 CrossRef CAS.
- N. W. S. Kam, T. C. Jessop, P. A. Wender and H. Dai, Nanotube molecular transporters: internalization of carbon nanotube—protein conjugates into mammalian cells, J. Am. Chem. Soc., 2004, 126, 12200–12201 CrossRef CAS.
- N. W. S. Kam and H. Dai, Carbon nanotubes as intracellular protein transporters: generality and biological functionality, J. Am. Chem. Soc., 2005, 127, 6021–6026 CrossRef CAS.
- N. S. W. Kam, Z. Liu and H. Dai, Carbon nanotubes as intracellular transporters for proteins and DNA: an investigation of the uptake mechanism and pathway, Angew. Chem., Int. Ed., 2006, 46, 577–581 CrossRef.
- N. Jia, Q. Lian, H. Shen, C. Wang, X. Li and Z. Yang, Intracellular Delivery of Quantum Dots Tagged Antisense Oligodeoxynucleotides by Functionalized Multiwalled Carbon Nanotubes, NanoLetters, 2007, 7, 2976–2980 CrossRef CAS.
- L. Cao, X. Wang, M. J. Meziani, F. Lu, H. Wang, P. G. Luo, Y. Lin, B. A. Harruff, L. M. Veca, D. Mrray, S. Y. Xie and Y. P. Sun, Carbon Dots for Multiphoton Bioimaging, J. Am. Chem. Soc., 2007, 129, 11318–11319 CrossRef CAS.
- D. Pantarotto, J. P. Briand, M. Prato and A. Bianco, Translocation of bioactive peptides across cell membranes by carbon nanotubes, Chem. Commun., 2004, 16–17 RSC.
- D. Pantarroto, R. Singh, D. McCarthy, M. Erhardt, J. P. Briand, M. Prato and K. Kostarelos, Functionalised nanotubes for plasmid DNA gene delivery, Angew. Chem., Int. Ed., 2004, 43, 5242–5246 CrossRef CAS.
- M. Geiser and B. Rothen-Rutishauser, et al. Ultrafine particles cross cellular membranes by nonphagocytic mechanisms in lungs and in cultured cells, Environ. Health Perspect., 2005, 113, 1555–60 CrossRef.
- N. A. Monteiro-Riviere and R. J. Nemanich, et al. Multi-walled carbon nanotube interactions with human epidermal keratinocytes, Toxicol. Lett., 2005, 155, 377–84 CrossRef CAS.
- S. Foley and C. Crowley, et al. Cellular localisation of a water-soluble fullerene derivative, Biochem. Biophys. Res. Commun., 2002, 294, 116–19 CrossRef CAS.
- L. W. Zhang and N. A. Monteiro-Riviere, Mechanisms of Quantum Dot NP Cellular Uptake, Toxicol. Sci., 2009, 110, 138–155 CrossRef CAS.
- M. Ricarda-Lorenz, V. Holzapfel, A. Musyanovych, K. Nothelfer, P. Walther, H. Frank, K. Landfester, H. Schrezenmeier and V. Mailander, Uptake of Functionalized, Fluorescent-labeled Polymeric Particles in Different Cell Lines and Stem Cells, Biomaterials, 2006, 27, 2820–2828 CrossRef CAS.
- J. G. Teeguarden and P. M. Hinderliter, et al. Particokinetics in vitro: dosimetry considerations for in vitro NP toxicity assessments, Toxicol. Sci., 2007, 95, 300–12 CAS.
- A. M. Schrand, L. Dai, J. J. Schlager, S. M. Hussain and E. Osawa, Differential Biocompatibility of Carbon Nanotubes and Nanodiamonds, Diamond Relat. Mater., 2007, 16, 2118–2123 CrossRef CAS.
- S. Yu, M. Kang, H. Chang, K. Chen and Y. Yu, Bright Fluorescent Nanodiamonds: No Photobleaching and Low Cytotoxicity, J. Am. Chem. Soc., 2005, 127, 17604–17605 CrossRef CAS.
- A. M. Schrand, H. Huang, C. Carlson, J. J. Schlager, E. Ōsawa, S. M. Hussain and L. Dai, Are Diamond NPs Cytotoxic?, J.Phys. Chem. B Letters, 2007b, 111, 2–7 Search PubMed.
- Y. R. Chang, H. Y. Lee, K. Chen, C. C. Chang, D. S. Tsai, C. C. Fu, T. S. Lim, Y. K. Tzeng, C. Y. Fang, C. C. Han, H. C. Chang and W. Fann, Mass Production and Dynamic Imaging of Fluorescent NDs, Nature Technology, 2008, 3, 284–288 Search PubMed.
- A. Magrez, S. Kasas, V. Salicio, N. Pasquier, J. Seo, M. Celio, S. Catsicas, B. Schwaller and L. Forro, Cellular Toxicity of Carbon-Based Nanomaterials, Nano Lett., 2006, 6, 1121–1125 CrossRef CAS.
- H. Dumortier, S. Lacotte, G. Pastorin, R. Marega, W. Wu, D. Bonifazi, J.-P. Brian, M. Prato, S. Muller and A. Bianco, Functionalized Carbon Nanotubes are Non-toxic and preserve the Functionality of primary immune Cells, Nano Lett., 2006, 6, 1522–1528 CrossRef CAS.
- M. Shim, N. W. Shi Kam, R. J. Chen, Y. Li and H. Dai, Functionalization of Carbon Nanotubes for Biocompatibility and Biomolecular Recognition, Nano Lett., 2002, 2, 285–288 CrossRef CAS.
- R. Hardman, A toxicologic review of quantum dots: toxicity depends on physicochemical and environmental factors, Environ. Health Perspect., 2006, 114, 165–72 CrossRef.
- N. Gibson, O. Shenderova, A. Puzyr, K. Purtov, V. Grichko, T. J. M. Luo, Z. Fitzgerald, V. Bondar and D. Brenner, NDs for Detoxification, Tech Proc. of the 2007 NanoTech. Conf. and Trade Show, 2007, 2, 713–716 Search PubMed.
- Y. Yuan, Y. Chen, J.-H. Liu, H. Wang and Y. Liu, Biodistribution and Fate of NDs in Vivo, Diamond Relat. Mater., 2009, 18, 95–100 CrossRef CAS.
- V. Yu. Dolmatov, Application of Detonation Nanodiamond, in Ultra Nanocrystalline Diamond: Synthesis, Properties, and Applications, ed. O. Shenderova and D. Gruen, William Andrew Publishing, Norwich, NY, USA, 2006, 513–527 Search PubMed.
- H. Huang, E. Pierstorff, E. Ōsawa and D. Ho, Active Nanodiamond Hydrogels for Chemotherapeutic Delivery, Nano Lett., 2007, 7, 3305–3314 CrossRef CAS.
- B. R. Smith, M. Niebert, T. Plakhotnik and A. V. Zvyagin, Transfection and Imaging of Diamond Nanocrystals as Scattering Optical Labels, J. Lumin., 2007, 260–263 CrossRef CAS.
- C. Y. Cheng, E. Perevedentseva, J. S. Tu, P. H. Chung, C. L. Cheng, K. K. Liu, J. I. Chao, P. H. Chen and C. C. Chang, Direct and In Vitro Observation of Growth Hormone Receptor Molecules in A549 Human Lung Epithelial Cells by Nanodiamond Labeling, Appl. Phys. Lett., 2007, 90, 163903 CrossRef.
- F. Treussart, V. Jacques, E. Wu, T. Gacoin, P. Grangier and J. F. Roch, Photoluminescence of Single Colour Defects in 50nm Diamond Nanocrystals, Phys. B, 2006, 376–377, 926–929 CrossRef CAS.
- C. C. Fu, H. Y. Lee, K. Chen, T. S. Lim, H. Y. Wu, P. K. Lin, P. K. Wei, P. H. Tsao, H. C. Chang and W. Fann, Characterization and Application of Single Fluorescent NDs as Cellular Biomarkers, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 727–732 CrossRef CAS.
- J. R. Rabeau, A. Stacey, A. Rabeau, S. Prawer, F. Jelezko, I. Mirza and J. Wrachtrup, Single Nitrogen Vacancy Centers in Chemical Vapor Deposited Diamond Nanocrystals, Nano Lett., 2007, 7, 3433–3437 CrossRef CAS.
- K. Y. Han, K. I. Willig, E. Rittweger, F. Jelezko, C. Eggeling and S. W. Hell, Three-Dimensional Stimulated Emission Depletion Microscopy of Nitrogen-Vacancy Centers in Diamond Using Continuous-Wave Light, Nano Lett., 2009, 9, 3323–3329 CrossRef CAS.
- X. L. Kong, L. C. L. Huang, C. M. Hsu, W. H. Chen, C. C. Han and H. C. Chang, High Affinity Capture of Proteins by Diamond NPs for Mass Spectroscopic Analysis, Anal. Chem., 2005, 77, 259–265 CrossRef CAS.
- H. Huang, E. Pierstorff, E. Osawa and D. Ho, Protein-Mediated Assembly of Nanodiamond Hydrogels into a Biocompatible and Biofunctional Multilayer Nanofilm, ACS Nano, 2008, 2, 203–212 CrossRef CAS.
- L.-C. L. Huang and H.-C. Chang, Adsorption and Immobilization of Cytochrome c on NDs, Langmuir, 2004, 20, 5879–5884 CrossRef CAS.
- A. Kruger, Hard and soft: biofunctionalized diamond, Angew. Chem., Int. Ed., 2006, 45, 6426–7 CrossRef.
- A. Krueger, J. Stegk, L. Lu, Y. Liang and G. Jarre, Biotinylated Nanodiamond: Simple and Efficient Functionalization of Detonation Diamond, Langmuir, 2008, 24, 4200–04 CrossRef CAS.
- S. C. Hens, G. Cunningham, T. Tyler and O. Shenderova, Nanodiamond Bioconjugate Probes and their Collection by Electrophoresis, Diamond Relat. Mater., 2008, 17, 1858–1866 CrossRef CAS.
- A. Krueger, New Carbon Materials: Biological Applications of Functionalized Nanodiamond Materials, Chem.–Eur. J., 2008, 14, 1382–1390 CrossRef CAS.
- G. Oberdorster, E. Oberdorster and J. Oberdorster, Nanotoxicology: An Emerging Discipline Evolving from Studies of Ultrafine Particles, Environ. Health Perspect., 2005, 113(7), 823–839 CrossRef CAS.
- P. Hoet, I. Bruske-Hohlfeld and O. Salata, NPs: known and unknown health risks, J. Nanobiotechnol., 2004, 2, 12 CrossRef.
- A. Nel, T. Xia, L. Madler and N. Li, Toxic Potential of Materials at the Nanolevel, Science, 2006, 311, 622–627 CrossRef CAS.
- D. S. Blair, J. W. Rogers and C. H. F. Peden, Potassium-assisted, facile oxidation of Si3N4 thin films, J. Appl. Phys., 1990, 67, 2066–2073 CrossRef CAS.
- A. M. Schrand, L. K. Braydich-Stolle, J. J. Schlager, L. Dai and S. M. Hussain, Can Silver NPs be Useful as Potential Biological Labels?, Nanotechnology, 2008, 19, 1–13.
- A. M. Schrand, S. A. Ciftan Hens and O. A. Shenderova, Nanodiamond Particles: Properties and Perspectives for Bioapplications, Crit. Rev. Solid State Mat. Sci., 2009, 34, 18–74 Search PubMed.
- A. M. Schrand, J. J. Schlager, L. Dai and S. M. Hussain, Preparing Cells Dosed with Nanomaterials for Assessment of Internalization and Localization with Transmission Electron Microscopy, Nat. Protoc., 2010, 5, 744–757 Search PubMed.
- E. Macia, M. Ehrlich, R. Massol, E. Boucrot, C. Brunner and T. Kirchhausen, Dev. Cell, 2006, 10, 839–50 CrossRef CAS.
- A. J. Newton, T. Kirchhausen and V. N. Murthy, Proc. Natl. Acad. Sci. U. S. A., 2006, 103(47), 17955–17960 CrossRef CAS.
- J. Lippincott-Schwartz, L. Yuan, C. Tipper, M. Amherdt, L. Orci and R. D. Klausner, Cell, 1991, 67, 601–16 CrossRef CAS.
- T. Fujiwara, K. Oda, S. Yokota, A. Takatsuki and Y. Ikehara, J. Biol. Chem., 1988, 263(34), 18545–52 CAS.
- I. Gouzman, R. Brener and A. Hoffman, Surf. Sci., 1995, 333, 283 CrossRef.
- M. M. T. Khan and S. Srivastava, Polyhedron, 1988, 7, 1063 CrossRef CAS.
- C. D. Batich and D. S. Donald, J. Am. Chem. Soc., 1984, 106, 2758 CrossRef CAS.
- R. Nakagaki, D. C. Frost and C. A. McDowell, J. Electron Spectrosc. Relat. Phenom., 1981, 22, 289 CrossRef CAS.
- K. Xu and Q. Xue, A New Method for Deaggregation of Nanodiamond from Explosive Detonation: Graphitization-oxidation Method, Phys. Solid State, 2004, 46, 649–650 CrossRef CAS.
- E. Osawa, Recent progress and perspectives in single-digit Nanodiamond, Diamond Relat. Mater., 2007, 16, 2018–2022 CrossRef CAS.
- A. Krüger, F. Kataoka, M. Ozawa, T. Fujino, Y. Suzuki, A. E. Aleksenskii, A-Ya. Vul and E. Ōsawa, Unusually tight Aggregation in Detonation Nanodiamond: Identification and Disintegration, Carbon, 2005, 43, 1722–1730 CrossRef.
- A. Krueger, M. Ozawa, G. Jarre, Y. Liang, J. Stegk and L. Lu, Deagglomeration and Functionalisation of Detonation Diamond, Phys. Status Solidi A, 2007, 204, 2881–2887 CrossRef CAS.
- K. Xu and Q. Xue, Deaggregation of Ultradispersed Diamond from Explosive Detonation by a Graphitization-oxidation Method and by Hydroiodic Acid Treatment, Diamond Relat. Mater., 2007, 16, 277–282 CrossRef CAS.
- The Lysosome: Structure, Organization, and Function, in Fundamentals of Medical Cell Biology Vol. 4: Membranology and Subcellular Organelles, ed. E. Bittar, ch. 10, pp. 305–361 Search PubMed.
- O. Faklaris, V. Joshi, T. Irinopoulou, P. Tauc, M. Sennour, H. Girard, C. Gesset, J.-C. Arnault, A. Thorel and J.-P. Boudou, et al. Photoluminescent Diamond NPs for Cell Labeling: Study of the Uptake Mechanism in Mammalian Cells, ACS Nano, 2009, 3955–3962 CrossRef CAS.
- K. Ushizawa and Y. Sato, et al. Covalent immobilization of DNA on diamond and its verification by diffuse reflectance infrared spectroscopy, Chem. Phys. Lett., 2002, 351, 105–108 CrossRef CAS.
- V. Grichko, V. Grishko and O. Shenderova, Nanodiamond Bullets and Their Biological Targets, NanoBiotechnology, 2006, 2, 37–42 CrossRef CAS.
- N. Kossovsky and A. Gelman, et al. Surface-modified diamond NPs as antigen delivery vehicles, Bioconjugate Chem., 1995, 6, 507–11 CrossRef CAS.
- T. T. B. Nguyen and H. C. Chang, et al. Adsorption and hydrolytic activity of lysozyme on diamond nanocrystallites, Diamond Relat. Mater., 2007, 16, 872–876 CrossRef CAS.
- E. Perevedentseva and C. Y. Cheng, et al. The interaction of the protein lysozyme with bacteria E. coli observed using Nanodiamond labeling, Nanotechnology, 2007, 18, 1–7.
- K. K. Liu and F. Chen, et al. Alpha-bungarotoxin binding to target cell in a developing visual system by carboxylated Nanodiamond, Nanotechnology, 2008, 19, 1–10.
- M. Lewin, N. Carlesso, C. H. Tung, X. W. Tang, D. Cory, D. T. Scadden and R. Weissleder, Tat peptide-derivatized magnetic NPs allow in vivo tracking and recovery of progenitor cells, Nat. Biotechnol., 2000, 18, 410–414 CrossRef CAS.
- S. Santra, H. Yang, D. Dutta, J. T. Stanley, P. H. Holloway, W. Tan, B. M. Moudgil and R. A. Mericle, TAT Conjugated, FITC Doped Silica NPs for Bioimaging Applications, Chem. Commun., 2004, 2810–2811 RSC.
- G. Ruan, A. Agrawal, A. I. Marcus and S. Nie, Imaging and Tracking of Tat Peptide-conjugated Quantum Dots in Living Cells: New Insights into NP Uptake, Intracellular Transport, and Vesicle Shedding, J. Am. Chem. Soc., 2007, 129, 14759–14766 CrossRef CAS.
- C. W. Lu, Y. Hung, J. K. Hsiao, M. Yao, T. H. Chung, Y. S. Lin, S. H. Wu, S. C. Hsu, H. M. Liu, C. Y. Mou, C. S. Yang, D. M. Huang and Y. C. Chen, Bifunctional Magnetic Silica NPs for Highly Efficient Human Stem Cell Labeling, Nano Lett., 2007, 7, 149–154 CrossRef CAS.
- C. M. Sayes, A. A. Marchione, K. L. Reed and D. B. Warheit, Comparative Pulmonary Toxicity Assessments of C60 Water Suspensions in Rats: Few Differences in Fullerene Toxicity in Vivo in Contrast to in Vitro Profiles, Nano Lett., 2007, 7, 2399–2406 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2011 |
