Synthesis and NMDA receptor affinity of dexoxadrol analogues with modifications in position 4 of the piperidine ring†
Ashutosh
Banerjee
a,
Roland
Fröhlich
b,
Dirk
Schepmann
a and
Bernhard
Wünsch
*a
aInstitut für Pharmazeutische und Medizinische Chemie der Westfälischen Wilhems-Universität Münster, Hittorfstraße 58-62, D-48149, Münster, Germany. E-mail: wuensch@uni-muenster.de; Fax: +49-251-8332144; Tel: +49-251-8333311
bOrganisch-chemisches Institut der Westfälischen Wilhems-Universität Münster, Corrensstraße 40, D-48149, Münster, Germany
First published on 28th April 2010
Abstract
A series of dexoxadrol (3) analogues with various substituents at position 4 of the piperidine ring has been synthesized and pharmacologically evaluated. Key steps in the synthesis are a hetero-Diels–Alder reaction of the dioxolane-derived imine 10 with Danishefsky's diene 11 and replacement of the p-methoxybenzyl protective group by a Cbz group. All possible diastereomers were synthesized, respectively. It was shown that like-configuration of the ring junction (position 2 of the piperidine ring and position 4 of the dioxolane ring) and axial orientation of the C-4 substituent are crucial for high NMDA receptor affinity. 2-(2,2-Diphenyl-1,3-dioxolan-4-yl)piperidine with a hydroxy moiety at position 4 (17d, WMS-2508, Ki = 44 nM) represents the most potent NMDA antagonist with high selectivity against σ1 and σ2 receptors, and the polyamine binding site of the NMDA receptor. The high sensitivity of the receptor ligand interaction became evident since methyl ethers with unlike-configuration of the ring junction (19a, 19b) prefer the σ1 receptor with high selectivity.
Introduction
(S)-Glutamate, which is the main excitatory neurotransmitter in the central nervous system (CNS), and other excitatory amino acids (EAAs) operate through two different classes of glutamate receptors. The first class are ionotropic receptors, which consists of NMDA (N-methyl-D-aspartate), AMPA (2-amino-3-(3-hydroxy-5-methylisoxazol-4-yl)propionate) and kainate receptors, and the second class are metabotropic receptors with the mGluR1–mGluR8 subtypes.1 The NMDA receptor subtype, which is selectively activated by N-methyl-D-aspartate, represents the target system of the following report.The physiological activation of the NMDA receptor is important for the development of neurons and for processes like memory and learning. However, an overactivation of the NMDA receptor leads to an increased influx of Ca2+-ions into neurons resulting in damage of the neuronal cells (excitotoxicity). Therefore, compounds blocking the excessive influx of Ca2+-ions through the NMDA receptor associated ion channel into neurons are of major interest as neuroprotective agents, which may be used for the therapy of cerebral ischemia, stroke, epilepsy and trauma (brain injury). Moreover, permanent increased activation of NMDA receptors has been discussed to be involved in the development of chronic neurodegenerative disorders like Parkinson`s disease, Alzheimer`s disease and alcohol dependency. Altogether the NMDA receptor represents an interesting target for the development of novel drugs for the therapy of neurological disorders.2
The opening of the NMDA receptor-associated cation channel is controlled by various ligands interacting with different binding sites at the receptor protein. The receptor comprises binding sites for (S)-glutamate, glycine, polyamines, Zn2+, Mg2+, H+ and phencyclidine (PCP, 1, Fig. 1).3
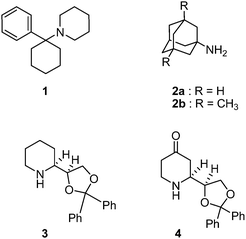 | ||
| Fig. 1 Structures of some important NMDA receptor antagonists: phencyclidine (PCP, 1); amantadine (2a); memantine (2b); dexoxadrol (3); 4-oxodexoxadrol (4). | ||
Our interest has been focused on the PCP binding site, which is located within the cation channel. Compounds interacting with the PCP binding site work as non-competitive NMDA receptor antagonists by blocking the cation channel and inhibiting the passage of cations, in particular the influx of Ca2+-ions. Up to now, memantine (2b, Fig. 1) is the only NMDA receptor antagonist with moderate NMDA receptor affinity (Ki = 1.2 μM),4 which is used for the treatment of severe Alzheimer's Disease.4 The particular advantage of memantine is the fast off rate kinetic preventing the drug from accumulating in the ion channel. Therefore, memantine leaves rapidly inactivated ion channels and is not present when the next normal impulse arrives. The physiological neurotransmission is not disturbed by memantine resulting in minimal adverse effects. In contrast, pathologically tonically activated NMDA receptors are blocked by memantine.4 Parkinson's Disease can be treated with the desmethyl derivative amantadine (2a).
In contrast to memantine (2b), the piperidine derivative dexoxadrol (3) binds with high affinity (Ki = 11.4 nM) at the phencyclidine binding site within the NMDA receptor associated cation channel. Dexoxadrol was synthesized by Hardie et al. in the 1960s and revealed local anesthetic, spasmolytic, and central nervous system activity.5 Later, phencyclidine-like dissociative anesthetic activities were found.6 Unfortunately, clinical trials of dexoxadrol had to be stopped because of the psychotomimetic side effects. Since the severe side effects of dexoxadrol are attributed to its high NMDA receptor affinity and unfavorable kinetic properties, novel analogues with moderate affinities between those of dexoxadrol and memantine should display the desired pharmacological activity with an improved side effect profile.
Several derivatives of dexoxadrol have been synthesized in order to get more insight into the structure–affinity relationships within this compound class. In particular, the substituents and the size and the heteroatoms of the oxygen heterocycle have been modified.7 In addition to various N-substituents, the piperidine ring has been exchanged for simple primary, secondary and tertiary amines.8,9
In order to broaden the structure–affinity relationships of this class of NMDA antagonist, we became interested in the influence of substituents in the piperidine moiety on the NMDA receptor affinity. For this purpose, a novel strategy for the synthesis of oxo-dexoxadrol analogues and homologues has been elaborated involving a hetero-Diels–Alder reaction.10,11 The resulting racemic oxo-dexoxadrol 4 with the same relative configuration as dexoxadrol had a NMDA receptor affinity of 470 nM.10 This promising affinity stimulated the synthesis of further dexoxadrol derivatives modified in position 4 of the piperidine moiety.
Herein we report on the synthesis and NMDA receptor affinity of all the possible diastereomeric dexoxadrol analogues bearing OH, OCH3, NH2 and NHCH3 groups at position 4 of the piperidine ring. Since the first experiments with alcohols were very promising, primary amines representing bioisosteres12 of the alcohols, methyl ethers, which cannot act as H-bond donors and methylamines with the same size as methyl ethers but having H-bond donor and H-bond acceptor properties were included into the study.
Chemistry
The synthesis was started with the acetalization of benzophenone (6) with glycerol (5) to yield the dioxolane 7.10,11 Swern oxidation13 of the primary alcohol 7 afforded the aldehyde 8,10 which was condensed with p-methoxybenzylamine (PMB-NH2, 9) to give the imine 10. In contrast to the reported synthesis with benzylamine,10,11 in this project the p-methoxybenzyl protective group was selected, because the chromatographic purification and separation of the products were easier. Without isolation the imine 10 was reacted with Danishefsky's diene 1114,15 in an Yb(OTf)3-catalyzed hetero-Diels–Alder reaction16 to afford the 1,2-disubstituted dihydropyridones 12a and 12c as a mixture of unlike- and like-configured diastereomers (unlike-12a![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) like12c = 43
like12c = 43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 57). Selective reduction of the double bond of 12a/12c with LiBEt3H in the presence of BF3·OEt2 yielded the piperidin-4-ones 13a (unlike configuration) and 13c (like configuration) in the ratio 41
57). Selective reduction of the double bond of 12a/12c with LiBEt3H in the presence of BF3·OEt2 yielded the piperidin-4-ones 13a (unlike configuration) and 13c (like configuration) in the ratio 41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 59, which were separated by flash chromatography (Scheme 1).
59, which were separated by flash chromatography (Scheme 1).
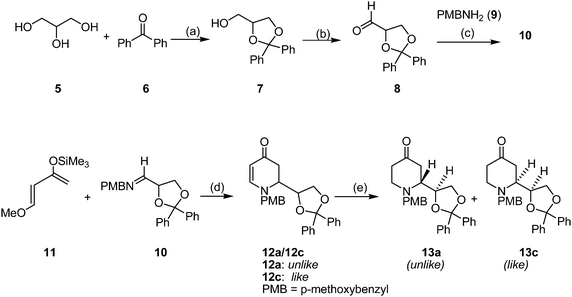 | ||
| Scheme 1 Synthesis of PMB-protected 4-oxo-dexoxadrol derivatives (only one enantiomer is shown, all compounds are racemic). Reagents and conditions: (a) TosOH·H2O, toluene, reflux, 6 h, 73%. (b) oxalyl chloride, DMSO, CH2Cl2, −78 °C, 30 min, then NEt3, rt, 72%. (c) 9, trimethyl orthoformate, rt, 16 h. (d) Yb(OTf)3, 11, THF, 0 °C, 4 h, then 25 °C, 16 h, 75% from 8. (e) BF3.OEt2, THF, −78 °C, 30 min, then LiBEt3H, −78 °C, 2 h, 37% (13a), 54% (13c). | ||
Reduction of 13a with NaBH4 resulted in the formation of a mixture (87![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 13) of diastereomeric alcohols, which could not be separated. The same problem was faced after NaBH4 reduction of the like-configured diastereomer 13c (diastereomeric ratio = 83
13) of diastereomeric alcohols, which could not be separated. The same problem was faced after NaBH4 reduction of the like-configured diastereomer 13c (diastereomeric ratio = 83![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 17). In order to improve the diastereoselectivity during the ketone reduction various reducing agents including L-Selectride (LiB(sec-Bu)3H), K-Selectride (KB(sec-Bu)3H), and LiBH4 were investigated, but the diastereoselectivity did not exceed 88
17). In order to improve the diastereoselectivity during the ketone reduction various reducing agents including L-Selectride (LiB(sec-Bu)3H), K-Selectride (KB(sec-Bu)3H), and LiBH4 were investigated, but the diastereoselectivity did not exceed 88![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12 (Table 1).
12 (Table 1).
Since the separation of the diastereomeric alcohols failed due to considerable tailing of the basic amines, the p-methoxybenzyl (PMB) protective group of 13a and 13c was replaced by the benzyloxycarbonyl (Cbz) protective group. After hydrogenolytic removal (H2, Pd/C) of the PMB group, the deprotected piperidines 14a and 14c were acylated with benzyl chloroformate (Cbz-Cl) in the presence of triethylamine to yield the Cbz-protected piperidines 15a and 15c, respectively. The ketones 15a and 15c were reduced unselectively with NaBH4 to afford 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixtures of diastereomeric alcohols 16a/16b and 16c/16d, respectively, which were separated by fc. Finally, the Cbz group of 16a–d was removed upon hydrogenolysis with H2, Pd/C to yield the piperidines 17a–d (Scheme 2).
1 mixtures of diastereomeric alcohols 16a/16b and 16c/16d, respectively, which were separated by fc. Finally, the Cbz group of 16a–d was removed upon hydrogenolysis with H2, Pd/C to yield the piperidines 17a–d (Scheme 2).
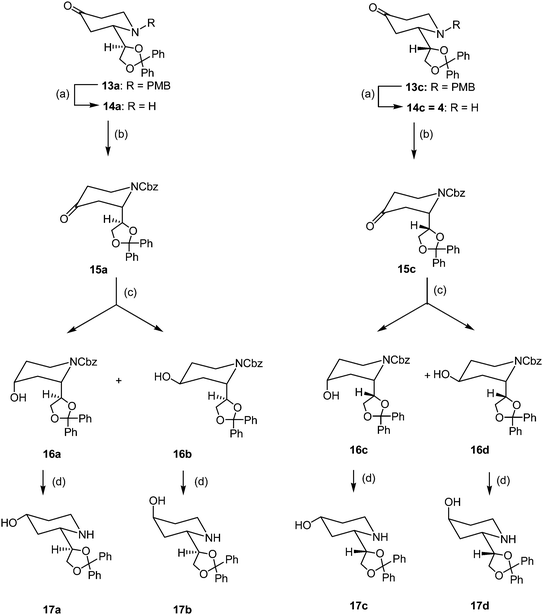 | ||
| Scheme 2 Synthesis of 4-hydroxy dexoxadrol derivatives (only one enantiomer is shown, all compounds are racemic). Reagents and conditions: (a) H2, Pd/C 10%, MeOH, 16 h, rt. (b) PhCH2OCOCl, NEt3, THF, 1 h, rt, 83% (15a from 13a), 86% (15c from 13c). (c) NaBH4, MeOH, 1 h, 0 °C, 43% (16a), 45% (16b), 43% (16c), 47% (16d). (d) H2, Pd/C 10%, MeOH, 4 h, rt, 80% (17a), 75% (17b), 88% (17c), 89% (17d). | ||
The relative configuration of the three centers of chirality was assigned by an X-ray crystal structure analysis. For this purpose, a sample of 17d was recrystallized from an n-hexane–CH2Cl2 mixture to get crystals, which were suitable for an X-ray structure analysis (Fig. 2). The structure of the racemic compound 17d proved the like-configuration (RS/RS) of the two centers of chirality at position 4 of the 1,3-dioxolane ring and position 2 of the piperidine ring (ring junction). The configuration of the third center of chirality at position 4 of the piperidine ring is correlating with the configuration of the other two centers of chirality. However, the piperidine 17d consists of the two enantiomers (R,R,R)-17d and (S,S,S)-17d. The relative configurations of the diastereomers of 17d as well as all precursors 12–16 were deduced from this X-ray crystal structure analysis and comparison of the corresponding NMR data.
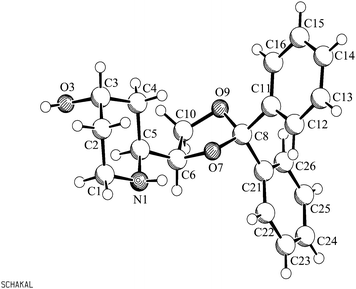 | ||
| Fig. 2 X-Ray crystal structure of 17d (racemic). | ||
In order to obtain the corresponding methoxy derivatives 19a–d the alcohols 16a–d were deprotonated with NaH and subsequently methylated with methyl iodide in DMF. Treatment of the methyl ethers 18a–d with H2 and Pd/C led to cleavage of the Cbz-protective group and the four diastereomeric methoxy derivatives 19a–d were isolated in 77–90% yields (Scheme 3).
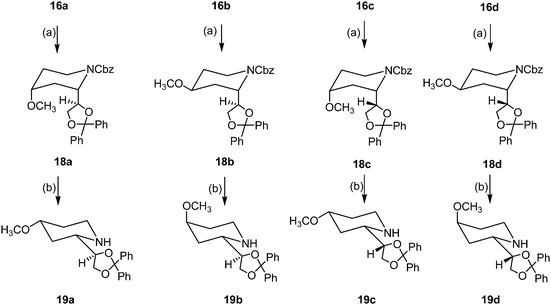 | ||
| Scheme 3 Synthesis of 4-methoxy dexoxadrol derivatives (only one enantiomer is shown, all compounds are racemic). Reagents and conditions: (a) NaH, MeI, DMF, 4 h, rt, 85% (18a), 80% (18b), 93% (18c), 91% (18d). (b) H2, Pd/C 10%, MeOH, 4 h, rt, 78% (19a), 90% (19b), 81% (19c), 77% (19d). | ||
During the analysis of the 1H NMR spectra an interesting flip of the piperidine ring was observed. Whereas the piperidine ring of derivatives with a proton (e.g.14, 17, 19) or a PMB group (e.g.13) attached to the N-atom adopts the 1C4 conformation, the corresponding Cbz derivatives (e.g.15, 16, 18) exist in the 1C4-conformation. Due to allylic strain17,18 the N-acyl-residue of 15, 16 and 18 forces the substituent in position 2 to adopt an axial orientation, which is only possible after changing the piperidine ring conformation. This piperidine ring flip is of particular importance for the interpretation of the 1H NMR data and the correct assignment of the relative configuration of all products. For example, the tt (J = 11.2/4.5 Hz) at 3.52 ppm in the 1H NMR spectrum of the Cbz-derivative 18d clearly indicates an axial orientation of 4-H, and the quintet (J = 2.9 Hz) at 3.60 ppm in the 1H NMR spectrum of the secondary amine 19d proves the equatorial position of 4-H.
For the synthesis of amino substituted dexoxadrol derivatives reductive aminations19 of the PMB-protected ketones 13a and 13c were performed. Although the transformations were successful, the separation of the formed diastereomeric amines was not possible. Therefore, again the Cbz protected ketones 15a and 15c were used for the introduction of amino moieties.
The reductive amination of unlike-configured ketone 15a with benzylamine or methylamine in the presence of NaBH(OAc)319 led to the diastereomeric benzylamines 20a/20b and methylamines 22a/22b in the ratio ≈7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, respectively, which were separated by flash chromatography (Scheme 4). Subsequent hydrogenolytic removal of the Cbz-protective group resulted in the primary amines 21a and 21b, since the benzyl moiety of the 4-benzylamino group was also cleaved, and the methylamines 23a and 23b.
3, respectively, which were separated by flash chromatography (Scheme 4). Subsequent hydrogenolytic removal of the Cbz-protective group resulted in the primary amines 21a and 21b, since the benzyl moiety of the 4-benzylamino group was also cleaved, and the methylamines 23a and 23b.
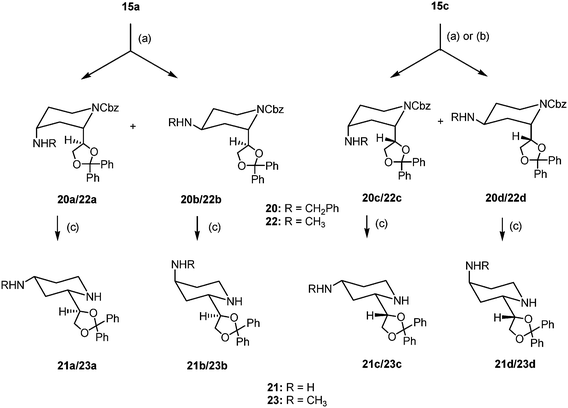 | ||
| Scheme 4 Synthesis of 4-amino dexoxadrol derivatives (only one enantiomer is shown, all compounds are racemic). Reagents and conditions: (a) PhCH2NH2 or MeNH2, NaBH(OAc)3, CH2Cl2, 2 h, rt, 58% (20a), 29% (20b), 92% (20c), 64% (22a), 26% (22b), 90% (22c). (b) PhCH2NH2 or MeNH2, trimethyl orthoformate, 14 h, rt; then NaBH4, MeOH, 1.5 h, 0 °C, 59% (20c), 37% (20d), 47% (22c), 36% (22d). (c) H2, Pd/C 10%, MeOH, 4 h, rt, 89% (21a), 87% (21b), 91% (21c), 83% (21d), 89% (23a), 78% (23b), 87% (23c), 82% (23d). | ||
In contrast to ketone 15a, a high diastereoselectivity was observed during the reductive amination of like-configured ketone 15c with benzylamine or methylamine and NaBH(OAc)3, which yielded the diastereomeric amines 20c/20d and 22c/22d in the ratio of about 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The main diastereomers 20c and 22c were isolated by flash chromatography, and subsequently treated with H2 and Pd/C to obtain the primary amine 21c and the methylamine 23c, respectively.
1. The main diastereomers 20c and 22c were isolated by flash chromatography, and subsequently treated with H2 and Pd/C to obtain the primary amine 21c and the methylamine 23c, respectively.
In order to get the fourth diastereomers 21d and 23d, ketone 15c was first converted into imines upon stirring with benzylamine or methylamine in trimethyl orthoformate overnight. After removal of the volatile components, methanol was added to the residue and the imines were reduced with NaBH4 at 0 °C. This procedure yielded the diastereomeric benzylamines 20c and 20d in the ratio 61![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 39 and the methylamines 22c and 22d in the ratio 57
39 and the methylamines 22c and 22d in the ratio 57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 43 after fc. After separation of the diastereomers hydrogenolytic cleavage led to the final amines 21d and 23d.
43 after fc. After separation of the diastereomers hydrogenolytic cleavage led to the final amines 21d and 23d.
During this reaction sequence the above described flip of the piperidine ring was also observed, when the N-substituent was changed from the Cbz-group (20/22) to a proton (21/23).
Receptor binding studies
The affinities of the novel dexoxadrol analogues to the PCP binding site of the NMDA receptor were determined in competition experiments using the potent and selective radioligand [3H]-(+)-MK-801. A membrane preparation from pig brain cortex was employed as receptor material.20,21 In addition to the affinity towards the PCP binding site the affinity of selected compounds towards the ifenprodil binding site of NR2B containing NMDA receptors was investigated. For this purpose the competitive receptor binding assay recently developed in our group was used. Briefly, in this assay tritium-labelled ifenprodil was employed as a radioligand. Membrane homogenates prepared from L(tk−) cells stably expressing recombinant human NR1a/NR2B receptors served as receptor material. The high density of NMDA receptors renders this system selective.22Since some potent NMDA receptor antagonists also interact with σ receptors and vice versa,23,24 the σ1 and σ2 receptor affinity of the piperidines was also determined. In the σ assays, the radioligands [3H]-(+)-pentazocine (σ1) and [3H]-(+)-ditolylguanidine (σ2), and membrane preparations from guinea pig brains (σ1) and rat livers (σ2) were used. In the σ2 assay, the σ1 binding sites were selectively masked with an excess of non-tritiated (+)-pentazocine.25,26,27
The Cbz-protected compounds 20 and 22 were also included into his study, because they contain a basic functional group. It should be noted that the basic amino group is at another position of the piperidine ring and the O-heterocycle in position 2 adopts an axial orientation in these compounds.
The receptor binding data are summarized in Table 2. For compounds with high NMDA affinity the competition curves with six compound concentrations were recorded three times and the resulting Ki-values with SEM are given. For compounds showing only low NMDA receptor interaction in the first screening, only the inhibition (%) at a compound concentration of 1 μM is given in Table 2. Some compounds indicated promising σ1 and/or σ2 affinities in the first screening. However, recording of the complete competition curves revealed only low affinity, and therefore the experiments were performed only once (n = 1).
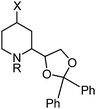
| Compound | X | R | Stereochemistryb | K i ± SEM/nMa | ||
|---|---|---|---|---|---|---|
| NMDA (PCP) ([3H]-(+)-MK-801) | σ1 ([3H]-(+)-pentazocine) | σ2([3H]-di-o-tolylguanidine) | ||||
| a The Ki-values were determined in three independent experiments (n = 3) unless otherwise noted. b All compounds were tested as racemic mixtures. In the column stereochemistry the relative configuration of the three centers of chirality is given. c Percent inhibition at a concentration of 1μM of the test compound. | ||||||
| 14a 10 | ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
H | 2S-4′R | 84% | 1390 ± 90 | 54% |
| 14c 10 | 2S-4′S | 470 ± 173 | 0% | 36% | ||
| 17a | OH | 2S,4R-4′R | 0%c | 318 (n = 1) | 0%c | |
| 17b | 2S,4S-4′R | 3%c | 259 (n = 1) | 30%c | ||
| 17c | 2S,4R-4′S | 527 ± 106 | 1250 (n = 1) | 2%c | ||
| 17d (WMS-2508) | 2S,4S-4′S | 44 ± 13 | 971 (n = 1) | 13%c | ||
| 19a | OCH3 | 2S,4R-4′R | 37%c | 17 ± 4.5 | 18%c | |
| 19b | 2S,4S-4′R | 21%c | 15 ± 1.8 | 23%c | ||
| 19c | 2S,4R-4′S | 49%c | 0%c | 5%c | ||
| 19d | 2S,4S-4′S | 981 ± 96 | 831 (n = 1) | 9%c | ||
| 20a | NHCH2Ph | Cbz | 2S,4R-4′R | 8%c | 12%c | 23%c |
| 20b | 2S,4S-4′R | 30%c | 20%c | 442 (n = 1) | ||
| 20c | 2S,4R-4′S | 20%c | 0%c | 14%c | ||
| 20d | 2S,4S-4′S | 15%c | 179 (n = 1) | 8%c | ||
| 21a | NH2 | H | 2S,4R-4′R | 26%c | 45%c | 15%c |
| 21b | 2S,4S-4′R | 25%c | 0%c | 13%c | ||
| 21c | 2S,4R-4′S | 44%c | 20%c | 26%c | ||
| 21d | 2S,4S-4′S | 17%c | 33%c | 17%c | ||
| 22a | NHCH3 | Cbz | 2S,4R-4′R | 7%c | 22%c | 28%c |
| 22b | 2S,4S-4′R | 7%c | 17%c | 18%c | ||
| 22c | 2S,4R-4′S | 10%c | 353 (n = 1) | 42%c | ||
| 22d | 2S,4S-4′S | 6%c | 8%c | 35%c | ||
| 23a | NHCH3 | H | 2S,4R-4′R | 16%c | 40%c | 44%c |
| 23b | 2S,4S-4′R | 16%c | 972 (n = 1) | 3670 (n = 1) | ||
| 23c | 2S,4R-4′S | 40%c | 21%c | 12%c | ||
| 23d | 2S,4S-4′S | 30%c | 32%c | 37%c | ||
| dexoxadrol | — | 11 ± 0.3 | — | — | ||
| (+)-MK-801 | — | 2.8 ± 1.8 | — | — | ||
| (+)-pentazocine | — | — | 4.2 ± 1.1 | — | ||
| di-o-tolyl guanidine | — | — | 89 ± 29 | 58 ± 18 | ||
The receptor binding data in Table 2 clearly show that the compounds 20–23 with a basic amino moiety at position 4 do not interact significantly with the PCP binding site of the NMDA receptor. Moreover, the σ1 and σ2 receptor affinity of the amines 20–23 was also very low. The Ki-value of the most potent basic σ1 ligand (20d) is considerably higher than 100 nM, indicating very low σ1 affinity.
In the alcohol (17) and methyl ether series (19) the stereochemistry plays a crucial role for the NMDA receptor affinity. In particular, the two centers of chirality at position 2 of the piperidine ring and in position 4 of the 1,3-dioxolane ring (ring junction) should be like-configured. This observation is in good accordance with literature results showing that dexoxadrol with (S,S)-configuration represents the most potent stereoisomer.6 Additionally, the stereocenter at position 4 of the piperidine ring should have the same configuration as the other two stereocenters. This results in an axial orientation of the functional group at position 4 of the piperidine ring. The effect of the stereochemistry on the NMDA receptor affinity becomes particularly evident for the alcohol series 17. Whereas 17a and 17b with unlike-configuration of the ring junction didn't interact considerably with the NMDA receptor, the like-configured derivatives 17c and 17d were considerably more active. The racemic piperidine 17d (WMS-2508) with an axially oriented OH-moiety provided a Ki-value of 44 nM, which is slightly increased compared with the Ki-value of the enantiomerically pure lead compound dexoxadrol (11.4 nM). Despite its 15-fold lower affinity, the potency of the diastereomer 17c (Ki = 527 nM) is still in a promising range considering the affinity of memantine (compare Introduction).
Replacement of the OH-moiety at position 4 of the piperidine derivative 17d by a methoxy group (19d) led to a drastic reduction of the NMDA receptor affinity (19d: Ki = 981 nM). We assume that the reduced affinity of the methyl ether 19d is due to the increased size of the methoxy group compared with the smaller hydroxy group.
Only negligible inhibition of radioligand binding at the ifenprodil binding site of the NMDA receptor was detected at a concentration of 1 μM for the test compounds 17c (30% inhibition), 17d (0% inhibition) and 19d (29% inhibition). The low inhibition indicates IC50 values higher than 1 μM for these test compounds. Obviously, at least the most potent ligand 17c, 17d and 19d reveal high selectivity for the PCP binding site over the ifenprodil binding site of the NMDA receptor.
Concerning σ1 receptor affinity, the stereochemistry at the ring junction is also very important. In contrast to the NMDA receptor, the σ1 receptor prefers piperidines with unlike-configuration of the ring junction. The unlike-configured alcohols 17a and 17b show moderate σ1 affinities and the corresponding methyl ethers 19a and 19b are very potent σ1 ligands (Ki = 17 and 15 nM). Comparison of the σ1 affinities of the diastereomers 17a/17b and 19a/19b clearly indicates that the configuration at position 4 of the piperidine moiety is of minor importance for the σ1 affinity.
Conclusion
The influence of substituents in position 4 of the piperidine moiety of dexoxadrol analogues on the NMDA and σ receptor affinity has been investigated. Potent NMDA antagonists were obtained, when a hydroxy moiety was introduced into position 4. like-configuration of the ring junction (position 2 of the piperidine ring and position 4 of the 1,3-dioxolane ring) as well as axial orientation of the OH moiety are crucial for high NMDA receptor affinity. The most potent NMDA antagonist 17d was very selective against σ1 receptors, σ2 receptors and the ifenprodil binding site of the NMDA receptor. However, changing of the configuration of the ring junction led to potent and selective σ1 ligands (19a, 19b). Since the prepared compounds are still racemic mixtures, the NMDA and σ1 receptor affinities will be further increased by enantiomerically pure ligands. Having the low NMDA receptor affinity of memantine with its ideal binding kinetic at the NMDA receptor in mind the alcohol 17c and the methyl ether 19d with moderate NMDA receptor affinity could also be of interest with respect to the side effect profile.Experimental
Chemistry general
Unless otherwise noted, moisture and oxygen sensitive reactions were conducted in dry glassware (Schlenk flask sealed with a rubber septum) under N2 (dried with phosphorous pentoxide (Granusic® A, Baker)). Yb(OTf)3 (Aldrich) was stored in a desiccator over P4O10 in vacuo at rt. Danishefsky's diene 11 was stored under N2 at −25 °C. THF was dried with sodium/benzophenone and was freshly distilled before use. Methanol was dried with magnesium and iodine, distilled, and stored over molecular sieves 4 Å. CH2Cl2 was dried with CaH2 and freshly distilled before use. Thin layer chromatography (tlc): Silica gel 60 F254 plates (Merck). Flash chromatography (fc): Silica gel 60, 40–64 μm (Merck); parentheses include: Diameter of the column (cm), length of the stationary phase (cm), eluent, fraction size (mL) and retention factor Rf. Melting point: melting point apparatus SMP 3 (Stuart Scientific), uncorrected. IR: IR spectrophotometer 480Plus FT-ATR-IR (Jasco). 1H NMR (400 MHz, 300 MHz, 600 MHz), 13C NMR (100 MHz) and 19F NMR (300 MHz, 600 MHz) spectra were recorded on a Unity Mercury Plus 400 (400 MHz) NMR spectrometer (Varian), Brucker AV 300 (300 MHz), and Varian Unity Plus 600 (600 MHz) operating at 23 °C. High temperature NMR was recorded in Unity Mercury Plus 400 (400 MHz) NMR spectrometer (Varian) at 100 °C. Chemical shifts δ are reported in parts per million (ppm) against the reference compound tetramethylsilane and calculated using the chemical shift of the signal of the residual non-deuterated solvent, for 19F NMR chemical shifts δ are reported in parts per million (ppm) against the reference compound CFCl3 and calculated using the chemical shift of the signal of the solvent. High temperature 1H NMR spectra were recorded using C6D5NO2 as solvent. MS: HRMS (ESI): Finnigan MAT 4200s, Brucker Daltonics Micro Tof and Waters Micromass Quatro LCZ, peaks are given in m/z (% of basis peak). EI, electron impact, MAT GCQ (Thermo-Finnigan). HPLC: Merck Hitachi Equipment; UV detector: L-7400; autosampler: L-7200; pump: L-7100; degasser: L-7614; column: LiChrospher® 60 RP-select B (5 μm); LiChroCART® 250–4 mm cartridge; flow rate: 1.000 mL min−1; injection volume: 5.0 μL; detection at λ = 210 nm; Method A: solvents: A: water with 0.05% (v/v) trifluoroacetic acid; B: acetonitrile with 0.05% (v/v) trifluoroacetic acid: gradient elution: (A%): 0–4 min: 90%, 4–29 min: gradient from 90%, to 0%, 29–31 min: 0%, 31–31.5 min: 0% to 90%, 31.5–40 min: 90%; Method B: solvents: A: water with 0.05% (v/v) trifluoroacetic acid; B: methanol with 0.05% (v/v) trifluoroacetic acid: gradient elution: (A%): 0–1 min: 80%, 1–22 min: gradient from 80%, to 0%, 22–30 min: 0%, 30–31.5 min: 0% to 80%, 31.5–40 min: 80%. The purity of all test compounds was greater than 95%, which was determined by one of the given HPLC methods.General procedures
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, 10 mL).
3, 10 mL).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, 10 mL).
3, 10 mL).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3, 20 mL).
3, 20 mL).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 for benzyl amine, EtOAc–NEtMe2 = 9
3 for benzyl amine, EtOAc–NEtMe2 = 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for methyl amine, 10 mL).
1 for methyl amine, 10 mL).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 for benzyl amine, EtOAc–NEtMe2 = 9
3 for benzyl amine, EtOAc–NEtMe2 = 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for methyl amine, 10 mL).
1 for methyl amine, 10 mL).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5
5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 10 mL) for the hydroxyl and methoxy derivatives and (2 cm, 12 cm, MeOH–NEtMe2 = 99
1, 10 mL) for the hydroxyl and methoxy derivatives and (2 cm, 12 cm, MeOH–NEtMe2 = 99![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 5 mL) for the amine derivatives.
1, 5 mL) for the amine derivatives.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 20 mL, Rf 0.20). Yellow oil, yield 1.0133 g (75%). IR (neat): ν/cm−1 = 1637 (m, C
1, 20 mL, Rf 0.20). Yellow oil, yield 1.0133 g (75%). IR (neat): ν/cm−1 = 1637 (m, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1579 (s, C
O), 1579 (s, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1071 (m, C–O–C). 1H NMR (CDCl3): δ (ppm) = 1.97 (d, J = 16.9 Hz, 0.43 H, 3x-H), 2.48 (dd, J = 16.9/2.8 Hz, 0.57 H, 3°-H), 2.68 (dd, J = 16.9/7.9 Hz, 0.57 H, 3°-H), 2.73 (dd, J = 16.9/7.3 Hz, 0.43 H, 3x-H), 3.45–3.49 (m, 0.43 H, 2x-H), 3.71 (dd, J = 8.6/5.9 Hz, 0.43 H, OCH2x), 3.77–3.79 (m, 0.57 H, OCH2°), 3.80 (s, 3 × 0.57 H, OCH3°), 3.81 (s, 3 × 0.43 H, OCH3x), 3.99 (dd, J = 8.3/6.9 Hz, 0.57 H, OCH2°), 4.04 (dd, J = 8.6/6.7 Hz, 0.43 H, OCH2x),4.05–4.09 (m, 0.57 H, OCH°), 4.29 (d, J = 14.9 Hz, 0.57 H, PhCH2°), 4.30–4.34 (m, 0.57 H, 2°-H), 4.41 (d, J = 14.9 Hz, 0.57 H, PhCH2°), 4.44 (d, J = 14.7 Hz, 0.43 H, PhCH2x), 4.65 (d, J = 14.9 Hz, 0.43 H, PhCH2x), 4.82 (dt, J = 9.8/6.3 Hz, 0.43 H, OCHx), 4.94 (d, J = 7.5 Hz, 1H, 5x-H + 5°-H), 6.85–6.89 (m, 3H, 2 × Ph + 6x-H + 6°-H), 7.02–7.15 (m, 3H, Ph), 7.26–7.34 (m, 5H, Ph), 7.47–7.52 (m, 4H, Ph). The ratio of 12a
C), 1071 (m, C–O–C). 1H NMR (CDCl3): δ (ppm) = 1.97 (d, J = 16.9 Hz, 0.43 H, 3x-H), 2.48 (dd, J = 16.9/2.8 Hz, 0.57 H, 3°-H), 2.68 (dd, J = 16.9/7.9 Hz, 0.57 H, 3°-H), 2.73 (dd, J = 16.9/7.3 Hz, 0.43 H, 3x-H), 3.45–3.49 (m, 0.43 H, 2x-H), 3.71 (dd, J = 8.6/5.9 Hz, 0.43 H, OCH2x), 3.77–3.79 (m, 0.57 H, OCH2°), 3.80 (s, 3 × 0.57 H, OCH3°), 3.81 (s, 3 × 0.43 H, OCH3x), 3.99 (dd, J = 8.3/6.9 Hz, 0.57 H, OCH2°), 4.04 (dd, J = 8.6/6.7 Hz, 0.43 H, OCH2x),4.05–4.09 (m, 0.57 H, OCH°), 4.29 (d, J = 14.9 Hz, 0.57 H, PhCH2°), 4.30–4.34 (m, 0.57 H, 2°-H), 4.41 (d, J = 14.9 Hz, 0.57 H, PhCH2°), 4.44 (d, J = 14.7 Hz, 0.43 H, PhCH2x), 4.65 (d, J = 14.9 Hz, 0.43 H, PhCH2x), 4.82 (dt, J = 9.8/6.3 Hz, 0.43 H, OCHx), 4.94 (d, J = 7.5 Hz, 1H, 5x-H + 5°-H), 6.85–6.89 (m, 3H, 2 × Ph + 6x-H + 6°-H), 7.02–7.15 (m, 3H, Ph), 7.26–7.34 (m, 5H, Ph), 7.47–7.52 (m, 4H, Ph). The ratio of 12a![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12c = 43
12c = 43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 57; ° = index for the major diastereomer 12c; x = index for the minor diastereomer 12a. 13C NMR (CDCl3): δ (ppm) = 36.7 (C-3°), 37.3 (C-3x), 55.2 (C-2°), 56.1 (C-2x), 58.2 (OC°H3, OCxH3), 58.7 (PhC°H2), 59.4 (PhCxH2), 66.7 (OC°H2), 66.8 (OCxH2), 74.1 (OC°H, OCxH), 96.8 (C5x), 98.5 (C5°), 109.4 (OC°O), 110.5 (OCxO), 114.2/ 114.3/ 125.6/ 125.8/ 125.9/ 126.0/ 128.0/ 128.1/ 128.2/ 128.3/ 128.5/ 128.7/ 128.9/ 141.6/ 141.8/ 141.9 (PhC), 152.6 (C-6x), 153.2 (C-6°), 159.4/159.5 (PhC), 188.7 (C-4x), 189.5 (C-4°). HRMS: calcd. for C28H27NO4H 442.2018, found 442.2013. HPLC (Method A): Purity 42% (12a), tR = 22.6 min, 53% (12c), tR = 22.0 min, together 95%.
57; ° = index for the major diastereomer 12c; x = index for the minor diastereomer 12a. 13C NMR (CDCl3): δ (ppm) = 36.7 (C-3°), 37.3 (C-3x), 55.2 (C-2°), 56.1 (C-2x), 58.2 (OC°H3, OCxH3), 58.7 (PhC°H2), 59.4 (PhCxH2), 66.7 (OC°H2), 66.8 (OCxH2), 74.1 (OC°H, OCxH), 96.8 (C5x), 98.5 (C5°), 109.4 (OC°O), 110.5 (OCxO), 114.2/ 114.3/ 125.6/ 125.8/ 125.9/ 126.0/ 128.0/ 128.1/ 128.2/ 128.3/ 128.5/ 128.7/ 128.9/ 141.6/ 141.8/ 141.9 (PhC), 152.6 (C-6x), 153.2 (C-6°), 159.4/159.5 (PhC), 188.7 (C-4x), 189.5 (C-4°). HRMS: calcd. for C28H27NO4H 442.2018, found 442.2013. HPLC (Method A): Purity 42% (12a), tR = 22.6 min, 53% (12c), tR = 22.0 min, together 95%.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 57, 1.0133 g, 2.3 mmol) was dissolved in THF (10 mL) and the solution was cooled down to −78 °C, BF3·OEt2 (0.58 mL, 4.6 mmol) was added slowly and the solution was stirred for 30 min at −78 °C. Then a 1M solution of LiEt3BH in THF (4.61 mL, 4.6 mmol) was added slowly and the reaction mixture was stirred for 2 h at −78 °C. Afterwards a saturated solution of NaHCO3 and EtOAc were added and the reaction mixture was warmed to rt. Then EtOAc was added, the organic layer was separated, the aqueous layer was extracted twice with EtOAc, the organic layer was dried (K2CO3), filtered and concentrated in vacuo. The residue was purified by fc (4 cm, 20 cm, petroleum ether–EtOAc = 85
57, 1.0133 g, 2.3 mmol) was dissolved in THF (10 mL) and the solution was cooled down to −78 °C, BF3·OEt2 (0.58 mL, 4.6 mmol) was added slowly and the solution was stirred for 30 min at −78 °C. Then a 1M solution of LiEt3BH in THF (4.61 mL, 4.6 mmol) was added slowly and the reaction mixture was stirred for 2 h at −78 °C. Afterwards a saturated solution of NaHCO3 and EtOAc were added and the reaction mixture was warmed to rt. Then EtOAc was added, the organic layer was separated, the aqueous layer was extracted twice with EtOAc, the organic layer was dried (K2CO3), filtered and concentrated in vacuo. The residue was purified by fc (4 cm, 20 cm, petroleum ether–EtOAc = 85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15, 20 mL).
15, 20 mL).
Compound 13a (Rf 0.16): Pale yellow oil, yield 0.3752 g (37%). IR (neat): ν/cm−1 = 1714 (s, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1205(s) / 1070 (s, –C–O–C–). 1H NMR (CDCl3): δ (ppm) = 2.21 (ddd, J = 14.7/ 5.0/1.3 Hz, 1H, 5-H), 2.26–2.33 (m, 1H, 5-H), 2.40–2.47 (m, 1H, 3-H), 2.66 (ddd, J = 14.8/5.9/0.9 Hz, 1H, 3-H), 2.78–2.84 (m, 1H, 6-H), 3.17 (ddd, J = 12.9/8.3/4.7 Hz, 1H, 6-H), 3.31 (q, J = 5.6 Hz, 1H, 2-H), 3.82 (s, 3H, OCH3), 3.85 (d, J = 13.4 Hz, 1H, PhCH2), 3.94 (t, J = 7.6 Hz, 1H, OCH2), 4.05 (d, J = 13.4 Hz, 1H, CH2Ph), 4.07 (dd, J = 7.8/7.0 Hz, 1H, OCH2), 4.37 (q, J = 7.2 Hz, 1H, OCH), 6.87–6.89 (m, 2H, Ph), 7.25–7.37 (m, 8H, Ph), 7.48–7.55 (m, 4H, Ph). HRMS: calculated for C28H29NO4H 444.2169, found 444.2164. EI (70 eV): m/z (rel. int.) = 443 (M+, 2), 366 (M-Ph, 2), 218 (M- diphenyldioxolanyl, 10), 121 (CH2C6H4OCH3, 100), 77 (Ph, 18). HPLC (Method A): Purity 95%, tR = 18.6 min.
O), 1205(s) / 1070 (s, –C–O–C–). 1H NMR (CDCl3): δ (ppm) = 2.21 (ddd, J = 14.7/ 5.0/1.3 Hz, 1H, 5-H), 2.26–2.33 (m, 1H, 5-H), 2.40–2.47 (m, 1H, 3-H), 2.66 (ddd, J = 14.8/5.9/0.9 Hz, 1H, 3-H), 2.78–2.84 (m, 1H, 6-H), 3.17 (ddd, J = 12.9/8.3/4.7 Hz, 1H, 6-H), 3.31 (q, J = 5.6 Hz, 1H, 2-H), 3.82 (s, 3H, OCH3), 3.85 (d, J = 13.4 Hz, 1H, PhCH2), 3.94 (t, J = 7.6 Hz, 1H, OCH2), 4.05 (d, J = 13.4 Hz, 1H, CH2Ph), 4.07 (dd, J = 7.8/7.0 Hz, 1H, OCH2), 4.37 (q, J = 7.2 Hz, 1H, OCH), 6.87–6.89 (m, 2H, Ph), 7.25–7.37 (m, 8H, Ph), 7.48–7.55 (m, 4H, Ph). HRMS: calculated for C28H29NO4H 444.2169, found 444.2164. EI (70 eV): m/z (rel. int.) = 443 (M+, 2), 366 (M-Ph, 2), 218 (M- diphenyldioxolanyl, 10), 121 (CH2C6H4OCH3, 100), 77 (Ph, 18). HPLC (Method A): Purity 95%, tR = 18.6 min.
Compound 13c (Rf 0.11): Pale yellow oil, yield 0.5477 g (54%). IR (neat): ν/cm−1 = 1709 (s, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1450 (m, C–H), 1205 (s) / 1069 (s, C–O–C). 1H NMR (CDCl3): δ (ppm) = 2.30 (dt, J = 15.1/3.8 Hz, 1H, 5-H), 2.51(ddd, J = 15.4/9.5/6.1 Hz, 1H, 5-H), 2.64 (dd, J = 14.9/5.9 Hz, 1H, 3-H), 2.71 (ddd, J = 14.8/4.0/1.2 Hz, 1H, 3-H), 2.96 (dt, J = 10.8/5.1 Hz, 1H, 6-H), 3.11–3.20 (m, 2H, 2-H + 6-H), 3.81 (s, 5H, OCH3, CH2Ph), 3.88 (dd, J = 8.0/7.0 Hz, 1H, OCH2) 4.08 (dd, J = 8.1/7.1, 1H, OCH2), 4.30 (q, J = 6.8 Hz, 1H, OCH), 6.87–6.89 (m, 2H, Ph), 7.23–7.37 (m, 8H, Ph), 7.43–7.45 (m, 2H, Ph), 7.52–7.54 (m, 2H, Ph). HRMS: calcd. for C28H29NO4H 444.2169, found 444.2173. EI (70 eV): m/z (rel. int.) = 443.2 (M+, 2), 218 (M-diphenyldioxolanyl, 12), 121(CH2C6H4OCH3, 100), 77 (Ph, 18). HPLC (Method A): Purity 96%, tR = 19.2 min.
O), 1450 (m, C–H), 1205 (s) / 1069 (s, C–O–C). 1H NMR (CDCl3): δ (ppm) = 2.30 (dt, J = 15.1/3.8 Hz, 1H, 5-H), 2.51(ddd, J = 15.4/9.5/6.1 Hz, 1H, 5-H), 2.64 (dd, J = 14.9/5.9 Hz, 1H, 3-H), 2.71 (ddd, J = 14.8/4.0/1.2 Hz, 1H, 3-H), 2.96 (dt, J = 10.8/5.1 Hz, 1H, 6-H), 3.11–3.20 (m, 2H, 2-H + 6-H), 3.81 (s, 5H, OCH3, CH2Ph), 3.88 (dd, J = 8.0/7.0 Hz, 1H, OCH2) 4.08 (dd, J = 8.1/7.1, 1H, OCH2), 4.30 (q, J = 6.8 Hz, 1H, OCH), 6.87–6.89 (m, 2H, Ph), 7.23–7.37 (m, 8H, Ph), 7.43–7.45 (m, 2H, Ph), 7.52–7.54 (m, 2H, Ph). HRMS: calcd. for C28H29NO4H 444.2169, found 444.2173. EI (70 eV): m/z (rel. int.) = 443.2 (M+, 2), 218 (M-diphenyldioxolanyl, 12), 121(CH2C6H4OCH3, 100), 77 (Ph, 18). HPLC (Method A): Purity 96%, tR = 19.2 min.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1206 (s) /1080 (s, C–O–C). 1H NMR (C6D5NO2, 100 °C): δ (ppm) = 1.92 (dd, J = 15.7/3.4 Hz, 1H, 3-H), 2.02–2.11 (m, 1H, 5-H), 2.15 (d broad, J = 15.7 Hz, 1H, 3-H), 2.34 (ddd, J = 15.2/7.5/2.6 Hz, 1H, 5-H), 3.13 (ddt, J = 15.1/11.4/3.9 Hz, 1H, 6-H), 3.56 (ddd, J = 15.0/ 7.5/3.5 Hz, 1H, 6-H), 3.60–3.64 (m, 1H, 2-H), 3.87–3.96 (m, 2H, OCH2Ph), 4.47–4.53 (m,1H, OCH), 4.75 (dd, J = 12.6/3.1 Hz, 1H, OCH2), 4.82 (dd, J = 12.6/3.2 Hz, 1H, OCH2), 6.76–7.17 (m, 15 H, Ph). HRMS: calcd. for C28H27NO5Na 480.1787, found 480.1781. HPLC (Method A): Purity 98%, tR = 22.9 min.
O), 1206 (s) /1080 (s, C–O–C). 1H NMR (C6D5NO2, 100 °C): δ (ppm) = 1.92 (dd, J = 15.7/3.4 Hz, 1H, 3-H), 2.02–2.11 (m, 1H, 5-H), 2.15 (d broad, J = 15.7 Hz, 1H, 3-H), 2.34 (ddd, J = 15.2/7.5/2.6 Hz, 1H, 5-H), 3.13 (ddt, J = 15.1/11.4/3.9 Hz, 1H, 6-H), 3.56 (ddd, J = 15.0/ 7.5/3.5 Hz, 1H, 6-H), 3.60–3.64 (m, 1H, 2-H), 3.87–3.96 (m, 2H, OCH2Ph), 4.47–4.53 (m,1H, OCH), 4.75 (dd, J = 12.6/3.1 Hz, 1H, OCH2), 4.82 (dd, J = 12.6/3.2 Hz, 1H, OCH2), 6.76–7.17 (m, 15 H, Ph). HRMS: calcd. for C28H27NO5Na 480.1787, found 480.1781. HPLC (Method A): Purity 98%, tR = 22.9 min.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1206(s)/ 1082 (s, C–O–C). 1H NMR (C6D5NO2, 100 °C): δ (ppm) = 2.03–2.17 (m, 1H, 3-H), 2.12 (ddd, J = 16.3/6.6/3.0 Hz, 1H, 5-H), 2.23 (ddd, J = 15.7/7.2/2.8 Hz, 1H, 5-H), 2.45 (dt, J = 15.8/2.8 Hz, 1H, 6-H), 3.18–3.26 (m, 1H, 3-H), 3.61 (ddd, J = 15.0/ 8.5/3.5 Hz, 1H, 6-H), 3.64–3.67 (m, 1H, 2-H), 3.93–4.04 (m, 2H, OCH2Ph), 4.31–4.36 (m,1H, OCH), 4.77 (dd, J = 12.6/2.8 Hz, 1H, OCH2), 4.82 (dd, J = 12.6/3.0 Hz, 1H, OCH2), 6.81–7.63 (m, 15 H, Ph). HRMS: calcd. for C28H27NO5Na 480.1787, found 480.1781. HPLC (Method A): Purity 98%, tR = 23.0 min.
O), 1206(s)/ 1082 (s, C–O–C). 1H NMR (C6D5NO2, 100 °C): δ (ppm) = 2.03–2.17 (m, 1H, 3-H), 2.12 (ddd, J = 16.3/6.6/3.0 Hz, 1H, 5-H), 2.23 (ddd, J = 15.7/7.2/2.8 Hz, 1H, 5-H), 2.45 (dt, J = 15.8/2.8 Hz, 1H, 6-H), 3.18–3.26 (m, 1H, 3-H), 3.61 (ddd, J = 15.0/ 8.5/3.5 Hz, 1H, 6-H), 3.64–3.67 (m, 1H, 2-H), 3.93–4.04 (m, 2H, OCH2Ph), 4.31–4.36 (m,1H, OCH), 4.77 (dd, J = 12.6/2.8 Hz, 1H, OCH2), 4.82 (dd, J = 12.6/3.0 Hz, 1H, OCH2), 6.81–7.63 (m, 15 H, Ph). HRMS: calcd. for C28H27NO5Na 480.1787, found 480.1781. HPLC (Method A): Purity 98%, tR = 23.0 min.
Compound 16a (Rf 0.13): Colorless oil, yield 0.118 g (43%). IR (neat): ν/cm−1 = 1670 (s, Cbz C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1204 (s)/1069 (s, C–O–C). 1H NMR (CDCl3): δ (ppm) = 1.54–1.57 (m broad, 3H, 3-H + 5-H), 1.63–1.68 (m broad, 1H, 3-H), 1.91–1.95 (m broad, 1H, 6-H), 3.23 (ddd, J = 14.1/11.9/4.4 Hz, 1H, 6-H), 3.75–3.80 (m broad, 1H, 2-H), 3.92 (s broad, 1H, OH), 3.98–4.02 (m, 2H, OCH2), 4.34–4.38 (m broad, 1H, 4-H), 4.46 (s broad, 1H, OCH), 4.98 (d, J = 12.1 Hz, 1H, OCH2Ph), 5.09 (d, J = 12.4 Hz, 1H, OCH2Ph), 7.17–7.27 (m, 10H, Ph), 7.36–7.42 (m, 5H, Ph). HRMS: calculated for C28H29NO5Na 482.1938, found 482.1938. HPLC (Method A): Purity 98%, tR = 22.19 min.
O), 1204 (s)/1069 (s, C–O–C). 1H NMR (CDCl3): δ (ppm) = 1.54–1.57 (m broad, 3H, 3-H + 5-H), 1.63–1.68 (m broad, 1H, 3-H), 1.91–1.95 (m broad, 1H, 6-H), 3.23 (ddd, J = 14.1/11.9/4.4 Hz, 1H, 6-H), 3.75–3.80 (m broad, 1H, 2-H), 3.92 (s broad, 1H, OH), 3.98–4.02 (m, 2H, OCH2), 4.34–4.38 (m broad, 1H, 4-H), 4.46 (s broad, 1H, OCH), 4.98 (d, J = 12.1 Hz, 1H, OCH2Ph), 5.09 (d, J = 12.4 Hz, 1H, OCH2Ph), 7.17–7.27 (m, 10H, Ph), 7.36–7.42 (m, 5H, Ph). HRMS: calculated for C28H29NO5Na 482.1938, found 482.1938. HPLC (Method A): Purity 98%, tR = 22.19 min.
Compound 16b (Rf 0.08): Colorless oil, yield 0.125 g (45%). IR (neat): ν/cm−1 = 1675 (s, Cbz C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1205 (s)/1070 (s, C–O–C). 1H NMR (C6D5NO2, 100 °C): δ (ppm) = 1.04 (qd, J = 12.6/4.8 Hz, 1H, 5-H), 1.29 (td, J = 12.5/6.3 Hz, 1H, 3-H), 1.53–1.59 (m broad, 2H, 3-H + 5-H), 1.81 (d, J = 4.7 Hz, 1H, OH), 2.83 (td, J = 13.5/2.6 Hz, 1H, 6-H), 3.49 (td, J = 7.7/2.2 Hz, 1H, 2-H), 3.71 (t broad, J = 7.3 Hz, 1H, OCH2), 3.76–3.83 (m, 1H, 4-H), 3.88 (d broad, J = 13.7 Hz, 1H, 6-H), 4.05 (q, J = 7.0 Hz, 1H, OCH), 4.31 (t broad, J = 6.5 Hz, 1H, OCH2), 4.69 (d, J = 12.7 Hz, 1H, OCH2Ph), 4.82 (d, J = 12.7 Hz, 1H, OCH2Ph), 6.78–7.63 (m, 15H, Ph). HRMS: calculated for C28H29NO5Na 482.1938, found 482.1934. HPLC (Method A): Purity 95%, tR = 21.6 min.
O), 1205 (s)/1070 (s, C–O–C). 1H NMR (C6D5NO2, 100 °C): δ (ppm) = 1.04 (qd, J = 12.6/4.8 Hz, 1H, 5-H), 1.29 (td, J = 12.5/6.3 Hz, 1H, 3-H), 1.53–1.59 (m broad, 2H, 3-H + 5-H), 1.81 (d, J = 4.7 Hz, 1H, OH), 2.83 (td, J = 13.5/2.6 Hz, 1H, 6-H), 3.49 (td, J = 7.7/2.2 Hz, 1H, 2-H), 3.71 (t broad, J = 7.3 Hz, 1H, OCH2), 3.76–3.83 (m, 1H, 4-H), 3.88 (d broad, J = 13.7 Hz, 1H, 6-H), 4.05 (q, J = 7.0 Hz, 1H, OCH), 4.31 (t broad, J = 6.5 Hz, 1H, OCH2), 4.69 (d, J = 12.7 Hz, 1H, OCH2Ph), 4.82 (d, J = 12.7 Hz, 1H, OCH2Ph), 6.78–7.63 (m, 15H, Ph). HRMS: calculated for C28H29NO5Na 482.1938, found 482.1934. HPLC (Method A): Purity 95%, tR = 21.6 min.
Compound 16c (Rf 0.26): Colorless oil, yield 78.9 mg (43%). IR (neat): ν/cm−1 = 1671 (s, Cbz C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1201(s) / 1049 (s, C–O–C). 1H NMR (CDCl3): δ (ppm) = 1.19 (s broad, 2H, 3-H), 1.65–1.56 (m, 3H, 5-H + OH), 2.12 (d broad, J = 15.0 Hz, 1H, 6-H), 3.06–3.37 (m, 1H, 6-H), 3.87–3.91 (m broad, 3H, 2-H + 2OCH2), 4.07 (s, 1H, 4-H), 4.13–4.77 (m, 1H, OCH), 5.00–5.07 (m, 2H, OCH2Ph), 6.90–7.69 (m, 15H, Ph). HRMS: calculated for C28H29NO5Na 482.1938, found 482.1935. HPLC (Method A): Purity 98%, tR = 22.2 min.
O), 1201(s) / 1049 (s, C–O–C). 1H NMR (CDCl3): δ (ppm) = 1.19 (s broad, 2H, 3-H), 1.65–1.56 (m, 3H, 5-H + OH), 2.12 (d broad, J = 15.0 Hz, 1H, 6-H), 3.06–3.37 (m, 1H, 6-H), 3.87–3.91 (m broad, 3H, 2-H + 2OCH2), 4.07 (s, 1H, 4-H), 4.13–4.77 (m, 1H, OCH), 5.00–5.07 (m, 2H, OCH2Ph), 6.90–7.69 (m, 15H, Ph). HRMS: calculated for C28H29NO5Na 482.1938, found 482.1935. HPLC (Method A): Purity 98%, tR = 22.2 min.
Compound 16d (Rf 0.16): Colorless oil, yield 87.5 mg (47%). IR (neat): ν/cm−1 = 1671 (s, Cbz C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1201 (s) / 1049 (s, C–O–C). 1H NMR (CDCl3): δ (ppm) = 1.15–1.23 (m, 3H, 3-H + 5-H), 1.83 (s broad, 1H, OH), 2.33 (d broad, J = 10.8 Hz, 1H, 5-H), 2.69–3.07 (m, 1H, 6-H), 3.89 (s broad, 2H, 6-H + 2-H), 3.95–4.05 (m, 1H, 4-H), 4.05–4.19 (m, 1H, OCH2), 4.19–4.27 (m, 1H, OCH2), 4.28–4.52 (m, 1H, OCH), 5.03 (s broad, 2H, OCH2Ph), 6.94–7.79 (m, 15H, Ph). HRMS: calculated for C28H29NO5Na 482.1938, found 482.1930. HPLC (Method A): Purity 96%, tR = 21.6 min.
O), 1201 (s) / 1049 (s, C–O–C). 1H NMR (CDCl3): δ (ppm) = 1.15–1.23 (m, 3H, 3-H + 5-H), 1.83 (s broad, 1H, OH), 2.33 (d broad, J = 10.8 Hz, 1H, 5-H), 2.69–3.07 (m, 1H, 6-H), 3.89 (s broad, 2H, 6-H + 2-H), 3.95–4.05 (m, 1H, 4-H), 4.05–4.19 (m, 1H, OCH2), 4.19–4.27 (m, 1H, OCH2), 4.28–4.52 (m, 1H, OCH), 5.03 (s broad, 2H, OCH2Ph), 6.94–7.79 (m, 15H, Ph). HRMS: calculated for C28H29NO5Na 482.1938, found 482.1930. HPLC (Method A): Purity 96%, tR = 21.6 min.
A sample of compound 17d was recrystallized with n-hexane–CH2Cl2 mixture and the resulting crystals were used for the X-ray crystal structure analysis.
Data set was collected with a Nonius KappaCCD diffractometer. Programs used: data collection COLLECT (Nonius B.V., 1998), data reduction Denzo-SMN (Z. Otwinowski and W. Minor, Methods in Enzymology, 1997, 276, 307–326), absorption correction Denzo (Z. Otwinowski, D. Borek, W. Majewski and W. Minor, Acta Crystallogr., 2003, A59, 228–234), structure solution SHELXS-97 (G. M. Sheldrick, Acta Crystallogr., 1990, A46, 467–473), structure refinement SHELXL-97 (G. M. Sheldrick, Acta Crystallogr., 2008, A64, 112–122), graphics SCHAKAL (E. Keller, Universität Freiburg, 1997).
CCDC 750839 contains the supplementary crystallographic data for this paper. These data can be obtained free of charge at http://www.ccdc.cam.ac.uk/conts/retrieving.html [or from the Cambridge Crystallographic Data Centre, 12 Union Road, Cambridge CB2 1EZ, UK; fax: (internet.) +44(1223)336-033, E-mail: mailto:deposit@ccdc.cam.ac.uk]
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1080 (s, O–CH3), 1205 (s) / 1027 (s, C–O–C). 1H NMR (CDCl3): δ (ppm) = 1.44–1.49 (m, 1H, 3-H), 1.51–1.53 (m, 1H, 5-H), 1.68 (ddd, J = 14.7/6.3/3.1 Hz, 1H, 3-H), 1.77 (d broad, J = 14.0 Hz, 1H, 5-H), 3.14 (s, 3H, OCH3), 3.20 (d broad, J = 13.2 Hz, 1H, 6-H), 3.39–3.41 (m, 1H, 2-H), 3.64 (t, J = 7.6 Hz, 1H, OCH2), 3.97 (d broad, J = 13.8 Hz, 1H, 6-H), 4.04 (t, J = 7.2 Hz, 1H, OCH2), 4.30–4.34 (m 1H, 4-H), 4.74 (dt, J = 9.5/6.9 Hz, 1H, OCH), 4.86 (s broad, 1H, OCH2Ph), 5.13 (d, J = 12.5 Hz, 1H, OCH2Ph), 7.12–7.42 (m, 15H, Ph). HRMS: calculated for C29H31NO5Na 496.2094, found 496.2099. HPLC (Method B): Purity 98%, tR = 21.5 min.
O), 1080 (s, O–CH3), 1205 (s) / 1027 (s, C–O–C). 1H NMR (CDCl3): δ (ppm) = 1.44–1.49 (m, 1H, 3-H), 1.51–1.53 (m, 1H, 5-H), 1.68 (ddd, J = 14.7/6.3/3.1 Hz, 1H, 3-H), 1.77 (d broad, J = 14.0 Hz, 1H, 5-H), 3.14 (s, 3H, OCH3), 3.20 (d broad, J = 13.2 Hz, 1H, 6-H), 3.39–3.41 (m, 1H, 2-H), 3.64 (t, J = 7.6 Hz, 1H, OCH2), 3.97 (d broad, J = 13.8 Hz, 1H, 6-H), 4.04 (t, J = 7.2 Hz, 1H, OCH2), 4.30–4.34 (m 1H, 4-H), 4.74 (dt, J = 9.5/6.9 Hz, 1H, OCH), 4.86 (s broad, 1H, OCH2Ph), 5.13 (d, J = 12.5 Hz, 1H, OCH2Ph), 7.12–7.42 (m, 15H, Ph). HRMS: calculated for C29H31NO5Na 496.2094, found 496.2099. HPLC (Method B): Purity 98%, tR = 21.5 min.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1092 (s, O–CH3), 1205(s) / 1026 (s, C–O–C). 1H NMR (C6D5NO2, 100 °C): δ (ppm) = 0.84–0.96 (m, 1H, 5-H), 1.18 (ddd, J = 13.4/4.7/2.5 Hz, 1H, 5-H), 1.55 (d broad, J = 13.2 Hz, 2H, 3-H), 2.70–2.81 (m,1H, 6-H), 2.84 (s, 3H, OCH3), 3.22–3.34 (m, 1H, 2-H), 3.49 (t, J = 7.8 Hz, 1H, OCH2), 3.69–3.73 (m,1H, OCH), 3.85 (d broad, J = 13.5 Hz, 1H, 6-H), 4.01–4.07 (m, 1H, OCH2), 4.26–4.30 (m, 1H, 4-H), 4.69 (d, J = 12.7 Hz, 1H, OCH2Ph), 4.82 (d, J = 12.7 Hz, 1H, OCH2Ph), 6.79–7.18 (m, 15H, -Ph). HRMS: calculated for C29H31NO5Na 496.2094, found 496.2090. HPLC (Method B): Purity 99%, tR = 21.2 min.
O), 1092 (s, O–CH3), 1205(s) / 1026 (s, C–O–C). 1H NMR (C6D5NO2, 100 °C): δ (ppm) = 0.84–0.96 (m, 1H, 5-H), 1.18 (ddd, J = 13.4/4.7/2.5 Hz, 1H, 5-H), 1.55 (d broad, J = 13.2 Hz, 2H, 3-H), 2.70–2.81 (m,1H, 6-H), 2.84 (s, 3H, OCH3), 3.22–3.34 (m, 1H, 2-H), 3.49 (t, J = 7.8 Hz, 1H, OCH2), 3.69–3.73 (m,1H, OCH), 3.85 (d broad, J = 13.5 Hz, 1H, 6-H), 4.01–4.07 (m, 1H, OCH2), 4.26–4.30 (m, 1H, 4-H), 4.69 (d, J = 12.7 Hz, 1H, OCH2Ph), 4.82 (d, J = 12.7 Hz, 1H, OCH2Ph), 6.79–7.18 (m, 15H, -Ph). HRMS: calculated for C29H31NO5Na 496.2094, found 496.2090. HPLC (Method B): Purity 99%, tR = 21.2 min.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1085 (s, O–CH3), 1205 (s) / 1027 (s, C–O–C). 1H NMR (CDCl3): δ (ppm) = 1.56 (ddd, J = 13.3/4.6/2.9 Hz, 1H, 3-H), 1.66–1.80 (m, 2H, 3-H + 5-H), 2.41 (d broad, J = 13.9 Hz, 1H, 5-H), 2.89–3.11 (m, 1H, 6-H), 3.37 (s, 3H, OCH3), 3.51–3.54 (m, 1H, 6-H), 3.84–3.85 (m broad, 3H, OCH2 + 2-H), 4.08–4.29 (m, 1H, 4-H), 4.62–4.84 (m, 1H, OCH), 5.04 (s, 2H, OCH2Ph), 6.80–7.63 (m, 15H, Ph). HRMS: calculated for C29H31NO5Na 496.2094, found 496.2093. HPLC (Method A): Purity 98%, tR = 25.16 min.
O), 1085 (s, O–CH3), 1205 (s) / 1027 (s, C–O–C). 1H NMR (CDCl3): δ (ppm) = 1.56 (ddd, J = 13.3/4.6/2.9 Hz, 1H, 3-H), 1.66–1.80 (m, 2H, 3-H + 5-H), 2.41 (d broad, J = 13.9 Hz, 1H, 5-H), 2.89–3.11 (m, 1H, 6-H), 3.37 (s, 3H, OCH3), 3.51–3.54 (m, 1H, 6-H), 3.84–3.85 (m broad, 3H, OCH2 + 2-H), 4.08–4.29 (m, 1H, 4-H), 4.62–4.84 (m, 1H, OCH), 5.04 (s, 2H, OCH2Ph), 6.80–7.63 (m, 15H, Ph). HRMS: calculated for C29H31NO5Na 496.2094, found 496.2093. HPLC (Method A): Purity 98%, tR = 25.16 min.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1092 (s, O–CH3), 1205 (s) / 1027 (s, C–O–C). 1H NMR (CDCl3): δ (ppm) = 1.22–1.27 (m, 1H, 3-H), 1.31 (s broad, 1H, 5-H), 1.82–2.01 (m, 1H, 3-H), 1.87–1.97 (m broad, 1H, 5-H), 2.72–3.06 (m, 1H, 6-H), 3.17 (s, 3H, OCH3), 3.52 (tt, J = 11.2/4.5 Hz, 1H, 4-H), 3.82–3.90 (m broad, 2H, 6-H + 2-H), 4.11–4.18 (m, 1H, OCH2), 4.25 (q, J = 6.7 Hz, 1H, OCH), 4.28–4.49 (m, 1H, OCH2), 5.04 (s, 2H, OCH2Ph), 6.84–7.67 (m, 15H, Ph). HRMS: calculated for C29H31NO5Na 496.2094, found 496.2093. HPLC (Method A): Purity 98%, tR = 24.48 min.
O), 1092 (s, O–CH3), 1205 (s) / 1027 (s, C–O–C). 1H NMR (CDCl3): δ (ppm) = 1.22–1.27 (m, 1H, 3-H), 1.31 (s broad, 1H, 5-H), 1.82–2.01 (m, 1H, 3-H), 1.87–1.97 (m broad, 1H, 5-H), 2.72–3.06 (m, 1H, 6-H), 3.17 (s, 3H, OCH3), 3.52 (tt, J = 11.2/4.5 Hz, 1H, 4-H), 3.82–3.90 (m broad, 2H, 6-H + 2-H), 4.11–4.18 (m, 1H, OCH2), 4.25 (q, J = 6.7 Hz, 1H, OCH), 4.28–4.49 (m, 1H, OCH2), 5.04 (s, 2H, OCH2Ph), 6.84–7.67 (m, 15H, Ph). HRMS: calculated for C29H31NO5Na 496.2094, found 496.2093. HPLC (Method A): Purity 98%, tR = 24.48 min.
Compound 20a (Rf 0.62): Colourless oil, yield 66.7 mg (58%). IR (neat): ν/cm−1 = 1690 (s, Cbz C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1205 (s)/1086 (s, C–O–C). 1H NMR (CDCl3): δ (ppm) = 1.27–1.32 (m, 1H, 3-H), 1.55–1.61 (m broad, 3H, 3-H + 5-H), 1.78–1.87 (m, 1H, 6-H), 1.93 (ddd, J = 14.0/6.0/3.9 Hz, 1H, 2-H), 2.97 (quint, J = 4.3 Hz, 1H, 4-H), 3.42 (td, J = 11.9/3.3 Hz, 1H, 6-H), 3.68 (d, J = 13.0 Hz, 1H, CH2Ph), 3.73 (d, J = 13.0 Hz, 1H, CH2Ph), 3.89 (t, J = 7.4 Hz, 1H, OCH2), 3.95 (s, 1H, NH), 4.05 (t, J = 7.4 Hz, 1H, OCH2), 4.36 (q, J = 6.8 Hz, 1H, OCH), 5.05 (d, J = 12.4 Hz, 1H, OCH2Ph), 5.22 (d, J = 12.5 Hz, 1H, OCH2Ph), 7.15–7.57 (m, 20H, Ph). HRMS: calculated for C35H36N2O4H 549.2748, found 549.2743. HPLC (Method A): Purity 98%, tR = 21.6 min.
O), 1205 (s)/1086 (s, C–O–C). 1H NMR (CDCl3): δ (ppm) = 1.27–1.32 (m, 1H, 3-H), 1.55–1.61 (m broad, 3H, 3-H + 5-H), 1.78–1.87 (m, 1H, 6-H), 1.93 (ddd, J = 14.0/6.0/3.9 Hz, 1H, 2-H), 2.97 (quint, J = 4.3 Hz, 1H, 4-H), 3.42 (td, J = 11.9/3.3 Hz, 1H, 6-H), 3.68 (d, J = 13.0 Hz, 1H, CH2Ph), 3.73 (d, J = 13.0 Hz, 1H, CH2Ph), 3.89 (t, J = 7.4 Hz, 1H, OCH2), 3.95 (s, 1H, NH), 4.05 (t, J = 7.4 Hz, 1H, OCH2), 4.36 (q, J = 6.8 Hz, 1H, OCH), 5.05 (d, J = 12.4 Hz, 1H, OCH2Ph), 5.22 (d, J = 12.5 Hz, 1H, OCH2Ph), 7.15–7.57 (m, 20H, Ph). HRMS: calculated for C35H36N2O4H 549.2748, found 549.2743. HPLC (Method A): Purity 98%, tR = 21.6 min.
Compound 20b (Rf 0.25): Colourless oil, yield 33.5 mg (29%). IR (neat): ν/cm−1 = 1688 (s, Cbz C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1205 (s)/1070 (s, C–O–C). 1H NMR (CDCl3): δ (ppm) = 1.20–1.29 (m broad, 1H, 3-H), 1.51 (s broad, 3H, 3-H + 5-H + NH), 1.73–1.76 (m broad, 1H, 5-H), 1.91–1.96 (m broad, 1H, 6-H), 2.97–3.17 (m, 2H, 6-H + 4-H), 3.77–3.83 (m broad, 3H, 2-H + CH2Ph), 4.00 (s broad, 1H, OCH2), 4.27–4.30 (m broad, 2H, OCH + OCH2), 4.89(d broad, J = 10.5 Hz, 1H, OCH2Ph), 5.15 (d, J = 11.8 Hz, 1H, OCH2Ph), 7.22–7.45 (m, 20H, Ph). HRMS: calculated for C35H36N2O4H 549.2748, found 549.2743. HPLC (Method A): Purity 97%, tR = 20.4 min.
O), 1205 (s)/1070 (s, C–O–C). 1H NMR (CDCl3): δ (ppm) = 1.20–1.29 (m broad, 1H, 3-H), 1.51 (s broad, 3H, 3-H + 5-H + NH), 1.73–1.76 (m broad, 1H, 5-H), 1.91–1.96 (m broad, 1H, 6-H), 2.97–3.17 (m, 2H, 6-H + 4-H), 3.77–3.83 (m broad, 3H, 2-H + CH2Ph), 4.00 (s broad, 1H, OCH2), 4.27–4.30 (m broad, 2H, OCH + OCH2), 4.89(d broad, J = 10.5 Hz, 1H, OCH2Ph), 5.15 (d, J = 11.8 Hz, 1H, OCH2Ph), 7.22–7.45 (m, 20H, Ph). HRMS: calculated for C35H36N2O4H 549.2748, found 549.2743. HPLC (Method A): Purity 97%, tR = 20.4 min.
Compound 20c (Rf 0.52): Colourless oil, yield 90.6 mg (59%). IR (neat): ν/cm−1 = 1693 (s, Cbz C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1203 (s)/1087 (s, C–O–C). 1H NMR (CDCl3): δ (ppm) = 1.17–1.20 (m, 1H, 3-H), 1.31–1.36 (m, 1H, 3-H), 1.46–1.62 (m broad, 2H, 5-H), 1.69 (d broad, J = 13.0 Hz, 1H, 6-H), 2.07 (d broad, J = 14.3 Hz, 1H, 6-H), 2.97 (quint, J = 3.7 Hz, 1H, 4-H), 3.20–3.29 (m broad, 1H, 2-H), 3.69 (d, J = 13.0 Hz, 1H, CH2Ph), 3.81 (d, J = 13.0 Hz, 1H, CH2Ph), 3.83–3.92 (m, 2H, NH + OCH2), 4.04–4.10 (m, 1H, OCH), 4.91–4.95 (m, 1H, OCH2), 4.98–5.07 (m, 2H, OCH2Ph), 7.08–7.26 (m, 15H, Ph), 7.32–7.38 (m, 5H, Ph). HRMS: calculated for C35H36N2O4H 549.2748, found 549.2743. HPLC (Method A): Purity 97%, tR = 22.6 min.
O), 1203 (s)/1087 (s, C–O–C). 1H NMR (CDCl3): δ (ppm) = 1.17–1.20 (m, 1H, 3-H), 1.31–1.36 (m, 1H, 3-H), 1.46–1.62 (m broad, 2H, 5-H), 1.69 (d broad, J = 13.0 Hz, 1H, 6-H), 2.07 (d broad, J = 14.3 Hz, 1H, 6-H), 2.97 (quint, J = 3.7 Hz, 1H, 4-H), 3.20–3.29 (m broad, 1H, 2-H), 3.69 (d, J = 13.0 Hz, 1H, CH2Ph), 3.81 (d, J = 13.0 Hz, 1H, CH2Ph), 3.83–3.92 (m, 2H, NH + OCH2), 4.04–4.10 (m, 1H, OCH), 4.91–4.95 (m, 1H, OCH2), 4.98–5.07 (m, 2H, OCH2Ph), 7.08–7.26 (m, 15H, Ph), 7.32–7.38 (m, 5H, Ph). HRMS: calculated for C35H36N2O4H 549.2748, found 549.2743. HPLC (Method A): Purity 97%, tR = 22.6 min.
Compound 20d (Rf 0.10): Colourless oil, yield 56.3 mg (37%). IR (neat): ν/cm−1 = 1697 (s, Cbz C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1204 (s)/1070 (s, C–O–C). 1H NMR (CDCl3): δ (ppm) = 1.21–1.28 (m broad, 2H, 3-H + NH), 1.31–1.39 (m broad, 2H, 3-H + 5-H), 1.84–1.91 (m broad, 1H, 5-H), 2.44 (d broad, J = 12.9 Hz, 1H, 6-H), 2.77–3.02 (m broad, 2H, 6-H + 4-H), 3.79 (d broad, J = 14.9 Hz, 2H, CH2Ph), 3.87–3.99 (m broad, 2H, 2-H + OCH2), 4.22–4.37 (m broad, 2H, OCH + OCH2), 5.11 (s broad, 2H, OCH2Ph), 7.27–7.47 (m, 20H, Ph). HRMS: calculated for C35H36N2O4H 549.2748, found 549.2741. HPLC (Method A): Purity 92%, tR = 22.0 min.
O), 1204 (s)/1070 (s, C–O–C). 1H NMR (CDCl3): δ (ppm) = 1.21–1.28 (m broad, 2H, 3-H + NH), 1.31–1.39 (m broad, 2H, 3-H + 5-H), 1.84–1.91 (m broad, 1H, 5-H), 2.44 (d broad, J = 12.9 Hz, 1H, 6-H), 2.77–3.02 (m broad, 2H, 6-H + 4-H), 3.79 (d broad, J = 14.9 Hz, 2H, CH2Ph), 3.87–3.99 (m broad, 2H, 2-H + OCH2), 4.22–4.37 (m broad, 2H, OCH + OCH2), 5.11 (s broad, 2H, OCH2Ph), 7.27–7.47 (m, 20H, Ph). HRMS: calculated for C35H36N2O4H 549.2748, found 549.2741. HPLC (Method A): Purity 92%, tR = 22.0 min.
Compound 22a (Rf 0.55): Colourless oil, yield 94.5 mg (64%). IR (neat): ν/cm−1 = 1688 (s, Cbz C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1207 (s)/1090 (s, C–O–C). 1H NMR (CDCl3): δ (ppm) = 1.25–1.31 (m, 1H, 3-H), 1.44–1.55 (m, 2H, 3-H + 5-H), 1.77–1.85 (m, 1H, 5-H), 1.89 (ddd, J = 13.6/6.1/3.8 Hz, 1H, 6-H), 2.34 (s, 3H, NCH3), 2.72 (quint, J = 5.0 Hz, 1H, 4-H), 3.37 (td, J = 12.8/3.8 Hz, 1H, 6-H), 3.85 (s broad, 1H, NH), 3.89 (t, J = 7.4 Hz, 1H, OCH2), 4.04 (t, J = 7.2 Hz, 1H, OCH2), 4.28 (q, J = 6.2 Hz, 1H, 2-H), 4.79 (q, J = 6.7 Hz, 1H, OCH), 5.01 (d, J = 12.5 Hz, 1H, OCH2Ph), 5.18 (d, J = 12.5 Hz, 1H, OCH2Ph), 7.23–7.35 (m, 10H, Ph), 7.47–7.53 (m, 5H, Ph). HRMS: calculated for C29H32N2O4H 473.2435, found 473.2437. HPLC (Method A): Purity 96%, tR = 19.2 min.
O), 1207 (s)/1090 (s, C–O–C). 1H NMR (CDCl3): δ (ppm) = 1.25–1.31 (m, 1H, 3-H), 1.44–1.55 (m, 2H, 3-H + 5-H), 1.77–1.85 (m, 1H, 5-H), 1.89 (ddd, J = 13.6/6.1/3.8 Hz, 1H, 6-H), 2.34 (s, 3H, NCH3), 2.72 (quint, J = 5.0 Hz, 1H, 4-H), 3.37 (td, J = 12.8/3.8 Hz, 1H, 6-H), 3.85 (s broad, 1H, NH), 3.89 (t, J = 7.4 Hz, 1H, OCH2), 4.04 (t, J = 7.2 Hz, 1H, OCH2), 4.28 (q, J = 6.2 Hz, 1H, 2-H), 4.79 (q, J = 6.7 Hz, 1H, OCH), 5.01 (d, J = 12.5 Hz, 1H, OCH2Ph), 5.18 (d, J = 12.5 Hz, 1H, OCH2Ph), 7.23–7.35 (m, 10H, Ph), 7.47–7.53 (m, 5H, Ph). HRMS: calculated for C29H32N2O4H 473.2435, found 473.2437. HPLC (Method A): Purity 96%, tR = 19.2 min.
Compound 22b (Rf 0.23): Colourless oil, yield 37.6 mg (26%). IR (neat): ν/cm−1 = 1693 (s, Cbz C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1208 (s)/1070 (s, C–O–C). 1H NMR (CDCl3): δ (ppm) = 1.15–1.17 (m broad, 1H, 3-H), 1.42–1.46 (m broad, 1H, 5-H), 1.73 (d broad, J = 11.1 Hz, 1H, 3-H), 1.91 (s broad, 2H, 5-H + NH), 2.42 (s, 3H, NCH3), 2.78 (s broad, 1H, 6-H), 2.93–3.10 (m broad, 2H, 6-H + 4-H), 3.85 (s broad, 1H, 2-H), 4.06 (t broad, J = 6.3 Hz, 1H, OCH2), 4.28–4.36 (m broad, 2H, OCH + OCH2), 4.87 (d, J = 11.6 Hz, 1H, OCH2Ph), 5.16 (d, J = 10.6 Hz, 1H, OCH2Ph), 7.22–7.36 (m, 10H, Ph), 7.44–7.51 (m, 5H, Ph). HRMS: calculated for C29H32N2O4H 473.2435, found 473.2430. HPLC (Method A): Purity 98%, tR = 19.7 min.
O), 1208 (s)/1070 (s, C–O–C). 1H NMR (CDCl3): δ (ppm) = 1.15–1.17 (m broad, 1H, 3-H), 1.42–1.46 (m broad, 1H, 5-H), 1.73 (d broad, J = 11.1 Hz, 1H, 3-H), 1.91 (s broad, 2H, 5-H + NH), 2.42 (s, 3H, NCH3), 2.78 (s broad, 1H, 6-H), 2.93–3.10 (m broad, 2H, 6-H + 4-H), 3.85 (s broad, 1H, 2-H), 4.06 (t broad, J = 6.3 Hz, 1H, OCH2), 4.28–4.36 (m broad, 2H, OCH + OCH2), 4.87 (d, J = 11.6 Hz, 1H, OCH2Ph), 5.16 (d, J = 10.6 Hz, 1H, OCH2Ph), 7.22–7.36 (m, 10H, Ph), 7.44–7.51 (m, 5H, Ph). HRMS: calculated for C29H32N2O4H 473.2435, found 473.2430. HPLC (Method A): Purity 98%, tR = 19.7 min.
Compound 22c (Rf 0.77): Colourless oil, yield 68.7 mg (47%). IR (neat): ν/cm−1 = 1692 (s, Cbz C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1205 (s)/1090 (s, C–O–C). 1H NMR (CDCl3): δ (ppm) = 1.18–1.21 (m, 1H, 3-H), 1.42 (d broad, J = 13.4 Hz, 1H, 5-H), 1.53–1.64 (m, 1H, 5-H), 1.68 (d broad, J = 13.7 Hz, 1H, 3-H), 1.95 (d broad, J = 14.2 Hz, 1H, 6-H), 2.34 (s, 3H, NCH3), 2.72 (quint, J = 4.0 Hz, 1H, 4-H), 3.20 (t broad, J = 12.7 Hz, 1H, 6-H), 3.70 (s broad, 1H, NH), 3.81–3.89 (m, 2H, OCH2), 3.96–4.02 (m, 1H, 2-H), 4.81 (dt, J = 9.5/5.9 Hz, 1H, OCH), 4.99 (d, J = 12.7 Hz, 1H, OCH2Ph), 5.04 (d, J = 12.7 Hz, 1H, OCH2Ph), 7.13–7.28 (m, 11H, Ph), 7.37–7.43 (m, 4H, Ph). HRMS: calculated for C29H32N2O4H 473.2435, found 473.2420. HPLC (Method A): Purity 96%, tR = 20.2 min.
O), 1205 (s)/1090 (s, C–O–C). 1H NMR (CDCl3): δ (ppm) = 1.18–1.21 (m, 1H, 3-H), 1.42 (d broad, J = 13.4 Hz, 1H, 5-H), 1.53–1.64 (m, 1H, 5-H), 1.68 (d broad, J = 13.7 Hz, 1H, 3-H), 1.95 (d broad, J = 14.2 Hz, 1H, 6-H), 2.34 (s, 3H, NCH3), 2.72 (quint, J = 4.0 Hz, 1H, 4-H), 3.20 (t broad, J = 12.7 Hz, 1H, 6-H), 3.70 (s broad, 1H, NH), 3.81–3.89 (m, 2H, OCH2), 3.96–4.02 (m, 1H, 2-H), 4.81 (dt, J = 9.5/5.9 Hz, 1H, OCH), 4.99 (d, J = 12.7 Hz, 1H, OCH2Ph), 5.04 (d, J = 12.7 Hz, 1H, OCH2Ph), 7.13–7.28 (m, 11H, Ph), 7.37–7.43 (m, 4H, Ph). HRMS: calculated for C29H32N2O4H 473.2435, found 473.2420. HPLC (Method A): Purity 96%, tR = 20.2 min.
Compound 22d (Rf 0.55): Colourless oil, yield 52.9 mg (36%). IR (neat): ν/cm−1 = 1693 (s, Cbz C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1206 (s)/1070 (s, C–O–C). 1H NMR (CDCl3): δ (ppm) = 1.28–1.38 (m broad, 1H, 3-H), 1.43–1.52 (m broad, 1H, 5-H), 1.94–2.02 (m broad, 1H, 3-H), 2.11–2.23 (m broad, 1H, 5-H), 2.35 (s, 3H, NCH3), 2.78–3.09 (m broad, 2H, 6-H + 4-H), 3.87–4.02 (m broad, 2H, 6-H + NH), 4.16–4.39 (m, 3H, 2-H + OCH + OCH2), 4.49 (t broad, J = 6.0 Hz, 1H, OCH2), 5.05–5.11 (m, 2H, OCH2Ph), 7.26–7.36 (m, 11H, Ph), 7.45–7.49 (m, 4H, Ph). HRMS: calculated for C29H32N2O4H 473.2435, found 473.2425. HPLC (Method A): Purity 94%, tR = 20.3 min.
O), 1206 (s)/1070 (s, C–O–C). 1H NMR (CDCl3): δ (ppm) = 1.28–1.38 (m broad, 1H, 3-H), 1.43–1.52 (m broad, 1H, 5-H), 1.94–2.02 (m broad, 1H, 3-H), 2.11–2.23 (m broad, 1H, 5-H), 2.35 (s, 3H, NCH3), 2.78–3.09 (m broad, 2H, 6-H + 4-H), 3.87–4.02 (m broad, 2H, 6-H + NH), 4.16–4.39 (m, 3H, 2-H + OCH + OCH2), 4.49 (t broad, J = 6.0 Hz, 1H, OCH2), 5.05–5.11 (m, 2H, OCH2Ph), 7.26–7.36 (m, 11H, Ph), 7.45–7.49 (m, 4H, Ph). HRMS: calculated for C29H32N2O4H 473.2435, found 473.2425. HPLC (Method A): Purity 94%, tR = 20.3 min.
Receptor binding studies
Materials and general procedures
Pig brains were a donation of the local slaughterhouse (Coesfeld, Germany). The guinea pig brains and rat livers were commercially available (Harlan-Winkelmann, Borchen, Germany). Homogenizer: Elvehjem Potter (B. Braun Biotech International, Melsungen, Germany). Centrifuge: high-speed cooling centrifuge model Sorvall RC-5C plus (Thermo Fisher Scientific, Langenselbold, Germany). Filter: printed Filtermat Typ A and B (Perkin Elmer LAS, Rodgau-Jügesheim, Germany), presoaked in 0.5% aqueous polyethylenimine for 2 h at room temperature before use. The filtration was carried out with a MicroBeta FilterMate-96 Harvester (Perkin Elmer). The scintillation analysis was performed using Meltilex (Typ A or B) solid scintillator (Perkin Elmer) and a MicroBeta Trilux scintillation analyzer (Perkin Elmer). The overall counting efficiency was 20%. All experiments were carried out in triplicates using standard 96-well-multiplates (Diagonal, Muenster, Germany). The IC50-values were determined in competition experiments with at least six concentrations of the test compounds and were calculated with the program GraphPad Prism® 3.0 (GraphPad Software, San Diego, CA, USA) by non-linear regression analysis. The Ki-values were calculated according to the formula of Cheng and Prusoff.28 The Ki-values are given as mean value ± SEM from three independent experiments.Affinity towards the phencyclidine binding site of the NMDA receptor, modified according to ref. 21
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 × g for 20 min at 4 °C. The pellet was resuspended in 5–6 volumes of TRIS/EDTA buffer (5 mM/1 mM, pH 7.5) and centrifuged again at 31
500 × g for 20 min at 4 °C. The pellet was resuspended in 5–6 volumes of TRIS/EDTA buffer (5 mM/1 mM, pH 7.5) and centrifuged again at 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g (20 min, 4 °C). This procedure was repeated twice. The final pellet was resuspended in 5–6 volumes of buffer, the protein concentration was determined according to the method of Bradford30 using bovine serum albumin as standard, and subsequently the preparation was frozen (−80 °C) in 1.5 mL portions containing about 0.8 mg protein mL−1.
000 × g (20 min, 4 °C). This procedure was repeated twice. The final pellet was resuspended in 5–6 volumes of buffer, the protein concentration was determined according to the method of Bradford30 using bovine serum albumin as standard, and subsequently the preparation was frozen (−80 °C) in 1.5 mL portions containing about 0.8 mg protein mL−1.
Affinity toards the σ1 receptor, modified according to ref. 25–27
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 g for 20 min at 4 °C. The pellet was resuspended in 5–6 volumes of buffer (50 mM Tris, pH 7.4) and centrifuged again at 23
500 g for 20 min at 4 °C. The pellet was resuspended in 5–6 volumes of buffer (50 mM Tris, pH 7.4) and centrifuged again at 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 g (20 min, 4 °C). This procedure was repeated twice. The final pellet was resuspended in 5–6 volumes of buffer, the protein concentration was determined according to the method of Bradford30 using bovine serum albumin as standard, and subsequently the preparation was frozen (−80 °C) in 1.5 mL portions containing about 1.5 mg protein mL−1.
500 g (20 min, 4 °C). This procedure was repeated twice. The final pellet was resuspended in 5–6 volumes of buffer, the protein concentration was determined according to the method of Bradford30 using bovine serum albumin as standard, and subsequently the preparation was frozen (−80 °C) in 1.5 mL portions containing about 1.5 mg protein mL−1.
Affinity toards the σ2 receptor (modified according to ref. 25–27)
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 20 min at 4 °C. The pellet was resuspended in buffer (50 mM Tris, pH 8.0) and incubated at room temperature for 30 min. After the incubation, the suspension was centrifuged again at 31
000 g for 20 min at 4 °C. The pellet was resuspended in buffer (50 mM Tris, pH 8.0) and incubated at room temperature for 30 min. After the incubation, the suspension was centrifuged again at 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 20 min at 4 °C. The final pellet was resuspended in buffer, the protein concentration was determined according to the method of Bradford30 using bovine serum albumin as standard, and subsequently the preparation was frozen (−80 °C) in 1.5 mL portions containing about 2 mg protein mL−1.
000 g for 20 min at 4 °C. The final pellet was resuspended in buffer, the protein concentration was determined according to the method of Bradford30 using bovine serum albumin as standard, and subsequently the preparation was frozen (−80 °C) in 1.5 mL portions containing about 2 mg protein mL−1.
Affinity towards the ifenprodil binding site of the NMDA receptor (NR2B affinity), modified according to ref. 22
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 ratio were used. The transformed L(tk−) cells were grown in Modified Earl's Medium (MEM) containing 10% of standardized FCS (Biochrom AG, Berlin, Germany). The expression of the NMDA receptor at the cell surface was induced after the cell density of the adherent growing cells had reached approximately 90% of confluency. For the induction, the original growth medium was replaced by growth medium containing 4 μM dexamethasone and 4 μM ketamine (final concentration). After 24 h the cells were harvested by scraping and pelleted (10 min, 5000 × g, Hettich Rotina 35R centrifuge, Tuttlingen, Germany).
5 ratio were used. The transformed L(tk−) cells were grown in Modified Earl's Medium (MEM) containing 10% of standardized FCS (Biochrom AG, Berlin, Germany). The expression of the NMDA receptor at the cell surface was induced after the cell density of the adherent growing cells had reached approximately 90% of confluency. For the induction, the original growth medium was replaced by growth medium containing 4 μM dexamethasone and 4 μM ketamine (final concentration). After 24 h the cells were harvested by scraping and pelleted (10 min, 5000 × g, Hettich Rotina 35R centrifuge, Tuttlingen, Germany).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g, 4 °C, Sorvall RC-5 plus, Thermo Scientific). The supernatant was discarded and the pellet resuspended in a defined volume of PBS yielding cell fragments of approximately 500
000 × g, 4 °C, Sorvall RC-5 plus, Thermo Scientific). The supernatant was discarded and the pellet resuspended in a defined volume of PBS yielding cell fragments of approximately 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells mL−1. The suspension of membrane homogenates was sonicated again (4 °C, 2 × 10 s cycles with a break of 10 min) and stored at −80 °C.
000 cells mL−1. The suspension of membrane homogenates was sonicated again (4 °C, 2 × 10 s cycles with a break of 10 min) and stored at −80 °C.
Acknowledgements
The NRW International Graduate School of Chemistry, Münster, which is funded by the Government of the State Nordrhein-Westfalen and the DAAD is deeply acknowledged for the financial support.References
- H. Bräuner-Osborne, J. Egebjerg, E. O. Nielsen, U. Madsen and P. Krogsgaard-Larsen, J. Med. Chem., 2000, 43, 2609–2645 CrossRef.
- W. P. Danysz, C. G. Bresink, I. Quack and G., Drug News Persp., 1995, 261–277 Search PubMed.
- D. Lodge, in Excitatory Amino Acid Receptors- Design of Agonists and Antagonists, ed. P. Krogsgaard-Larsen, J. J. Hansen, Ellis Harwood Ltd., Chichester, 1992, pp 201–215 Search PubMed.
- S. A. Lipton, Nat. Rev. Drug Discovery, 2006, 5, 160–170 CrossRef CAS.
- W. R. Hardie, J. Hidalgo, I. F. Halverstadt and R. E. Allen, J. Med. Chem., 1966, 9, 127–136 CrossRef CAS.
- A. E. Jacobson, E. A. Harrison, Jr., M. V. Mattson, M. F. Rafferty, K. C. Rice, J. H. Woods, G. Winger, R. E. Solomon, R. A. Lessor and J. V. Silverton, J. Pharmacol. Exp. Ther., 1987, 243, 110–117 CAS.
- A. Thurkauf, M. V. Mattson, S. Richardson, S. Mirsadeghi, P. L. Ornstein, E. A. Harrison, Jr., K. C. Rice, A. E. Jacobson and J. A. Monn, J. Med. Chem., 1992, 35, 1323–1329 CrossRef CAS.
- M. Aepkers and B. Wünsch, Bioorg. Med. Chem., 2005, 13, 6836–6849 CrossRef CAS.
- M. Sax and B. Wünsch, Curr. Top. Med. Chem., 2006, 6, 723–732 CrossRef CAS.
- M. Sax, K. Ebert, D. Schepmann, B. Wibbeling and B. Wünsch, Bioorg. Med. Chem., 2006, 14, 5955–5962 CrossRef CAS.
- M. Sax, R. Fröhlich, D. Schepmann and B. Wünsch, Eur. J. Org. Chem., 2008, 6015–6028 CrossRef CAS.
- G. A. Patani and E. J. LaVoie, Chem. Rev., 1996, 96, 3147–3176 CrossRef CAS.
- A. J. Mancuso and D. Swern, Synthesis, 1981, 165–185 CrossRef CAS.
- S. Danishefsky, Acc. Chem. Res., 1981, 14, 400–406 CrossRef CAS.
- L. Hansson and R. Carlson, Acta Chem. Scand., 1989, 43, 188–192 CrossRef CAS.
- P. Buonora, J. C. Olsen and T. Oh, Tetrahedron, 2001, 57, 6099–6138 CrossRef CAS.
- S. K. Malhotra and F. Johnson, J. Am. Chem. Soc., 1965, 87, 5493–5495 CrossRef CAS.
- E. E. Sugg, J. F. Griffin and P. S. Portoghese, J. Org. Chem., 1985, 50, 5032–5037 CrossRef CAS.
- A. F. Abdel-Magid and S. J. Mehrman, Org. Process Res. Dev., 2006, 10, 971–1031 Search PubMed.
- R. M. McKernan, S. Castro, J. A. Poat and E. H. F. Wong, J. Neurochem., 1989, 52, 777–785 CrossRef CAS.
- U. Wirt, D. Schepmann and B. Wünsch, Eur. J. Org. Chem., 2007, 462–475 CrossRef CAS.
- D. Schepmann, B. Frehland, K. Lehmkuhl, B. Tewes and B. Wünsch, J. Pharm. Biomed. Anal., 2010 DOI:10.1016/JPBA-D-09-01731R3.
- F. I. Carroll, P. Abraham, K. Parham, X. Bai, X. Zhang, G. A. Brine, S. W. Mascarella, B. R. Martin, E. L. May, C. Sauss, L. Di Paolo, P. Wallace, J. M. Walker and W. D. Bowen, J. Med. Chem., 1992, 35, 2812–2818 CrossRef CAS.
- E. L. May, M. D. Aceto, E. R. Bowman, C. Bentley, B. R. Martin, L. S. Harris, F. Medzihradsky, M. V. Mattson and A. E. Jacobson, J. Med. Chem., 1994, 37, 3408–3418 CrossRef CAS.
- C. A. Maier and B. Wünsch, J. Med. Chem., 2002, 45, 438–448 CrossRef CAS.
- C. A. Maier and B. Wünsch, J. Med. Chem., 2002, 45, 4923–4930 CrossRef CAS.
- R. Holl, D. Schepmann, R. Fröhlich, R. Grünert, P. J. Bednarski and B. Wünsch, J. Med. Chem., 2009, 52, 2126–2137 CrossRef CAS.
- Y.-C. Cheng and W. H. Prusoff, Biochem. Pharmacol., 1973, 22, 3099–3108 CrossRef CAS.
- Y. Ueda, H. Nakanishi and K. Yoshida, Life Sci., 1999, 65, 1477–1484 CrossRef CAS.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: HPLC data. See DOI: 10.1039/c0md00017e |
| This journal is © The Royal Society of Chemistry 2010 |
