A functionalized gold nanoparticles-assisted universal carrier for antisense DNA†
Jae-Hong
Kim‡
a,
Hyun Hye
Jang‡
b,
Sang-Mi
Ryou
ac,
Sudeok
Kim
b,
Jeehyeon
Bae
a,
Kangseok
Lee
*c and
Min Su
Han
*b
aGraduate School of Life Science and Biotechnology, CHA University, Seongnam 463-836, Republic of Korea
bDepartment of Chemistry, Chung-Ang University, Seoul 156-756, Republic of Korea. E-mail: mshan@cau.ac.kr
cDepartment of Life Science (BK21 Program), Chung-Ang University, Seoul 156-756, Republic of Korea. E-mail: kangseok@cau.ac.kr
First published on 10th May 2010
Abstract
In this study, single-stranded DNA functionalized gold nanoparticles (AuNP GDS) were proved to be efficient gene delivery systems for oligo antisense DNAs specific to any gene of interest without affecting normal cell physiology.
Gene therapy is attractive as a clinical treatment for cancers and genetic disorders of both congenital and acquired origin.1 Efficient gene delivery systems are central to the clinical treatment of genetic disorders and cancer and have attracted considerable attention in recent years.1 The use of recombinant viruses as gene carriers was the focus of early studies due to their high transfection efficiencies and levels of protein expression.2 However, these systems are critically limited because viral proteins trigger strong immune responses, which has resulted in a lack of FDA-approved products.3 Additionally, viral delivery systems are limited in scale-up procedures.4 As a result, numerous nonviral gene delivery systems such as cationic lipids, polymers, dendrimers, and peptides have been developed.5 However, nonviral gene delivery systems exhibit significantly reduced transfection efficiencies compared to viral systems due to numerous extra- and intracellular obstacles. Therefore, many researchers continue to focus on designing safe and efficient nonviral delivery vectors.
Recently, nanomaterials, including carbon nanotubes, iron oxide, silica, and gold nanoparticles, have been intensively studied as alternative nonviral gene delivery systems.6 Gold nanoparticles are attractive scaffolds for the creation of gene delivery systems because they are bioinert, nontoxic, and easily synthesized and functionalized.7 Thus far, several strategies for gene delivery systems have been developed, including mixed monolayer protected gold nanoparticles, complexes of polymer and gold nanoparticles, double-stranded DNA functionalized gold nanoparticles, and single stranded DNA functionalized gold nanoparticles.8 Single-stranded DNA functionalized gold nanoparticles were proved as a good gene delivery system and antisense.8f Moreover, these nanoparticles showed greater knockdown of gene expression, higher binding affinity for target DNA, higher immunity to nuclease, and lower cell toxicity than antisense DNA delivered by either Lipofectamine or Cytofectin. However, the system could deliver only antisense DNA covalently cross-linked to gold nanoparticles, which needed to be individually synthesized for each gene of interest, procedures that are both time consuming and inconvenient. Thus we developed a method that can be used for delivery of any antisense DNA oligo without the need for the synthesis of the gold nanoparticles covalently linked to antisense DNA oligo specific to the gene of interest.
In this paper, we describe the development of a gene delivery system showing the use of single-stranded DNA functionalized gold nanoparticles (henceforth referred to as “gold nanoparticle gene delivery system (AuNP GDS)”) as a general platform for loading and delivering antisense DNA specific to any gene of interest (Fig. 1). We used an oligonucleotide (oligo) bearing a partial sequence of RNA I (bases 13 to 30), which is involved in the replication of ColE1-type plasmid in Escherichia coli.9 The RNA I oligo was used because human cells do not contain nucleic acids complementary to the sequence of RNA I oligo used here and, therefore, it is not expected that introduction of RNA I oligo into the human cells interferes with expression of any human genes. AuNP GDS was prepared by functionalizing 13 nm gold nanoparticles with thiolated RNA I oligos.10 To test whether the AuNP GDS could be used as a gene delivery system, RNA I oligo-functionalized gold nanoparticles were annealed with antisense DNA to the p53 gene, and the resulting conjugates (AuNP GDS-AS) were applied to HeLa cells. The gene p53 is a well characterized transcription factor that plays a crucial role in the maintenance of cell-cycle arrest after DNA damage and in apoptosis as a tumor suppressor.11 Antisense DNAs to the p53 gene contained a sequence complementary to AuNP GDS followed by a 20 nucleotide-sequence complementary to an internal coding region of p53 (bases 886 to 905) for AS-p53-1.12 To verify the delivery of the antisense oligo into the cell, we used a 3′-end FITC-labeled oligo for producing AuNP GDS-p53-AS-FITC conjugates. Fluorescence microscopic analyses showed the presence of fluorescence from p53-AS-FITC in every cell tested (Fig. 2), indicating an excellent transfection efficiency of AuNP GDS-p53-AS-FITC. Previously, it was reported that single-stranded DNA functionalized gold nanoparticles readily enter cells.8f Our results show that AuNP GDS-AS conjugates containing partially double-stranded (18 base-pairs) DNA also enter cells with nearly 100% efficiency. It is possible that the efficient penetration of nanoparticles to cells was mediated with proteins in culture media.13
 | ||
| Fig. 1 Schematic of a single-stranded DNA functionalized gold nanoparticle-antisense. | ||
 | ||
| Fig. 2 Representative confocal fluorescence microscopy images of HeLa cells. (A) The cell nucleus was stained using DAPI. (B) HeLa cells were exposed to 1.0 nM AuNP GDS-p53-AS-FITC conjugates. (C) Transmission image of HeLa cells. (D) Overlay image of images A, B, and C. | ||
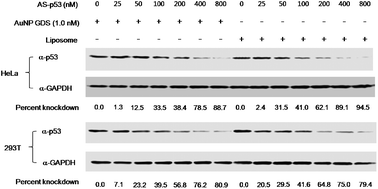
Next, we tested the ability of the antisense oligo to knockdown target gene expression. AuNP GDS-p53-AS conjugates were applied to HeLa cells and protein samples were prepared from the cultures 24 h after transfection for quantitative analysis of p53 using Western blot analysis. Relative expression levels of p53 protein were quantified using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as a reference protein. As shown in Fig. 3, AuNP GDS-p53-AS conjugates efficiently knocked down p53 expression in a p53 antisense DNA concentration dependent manner. When p53-AS was delivered into cells using a liposome-based reagent (Lipofectamine 2000, Invitrogen), the knockdown efficiency was as efficient as that of AuNP GDS. In addition, AuNP GDS itself did not affect expression levels of target proteins or the reference protein. We tested two other human cell lines, 293T (Fig. 3) and K562 (Supplementary Fig. S1†) for transfection of p53-AS annealed to AuNP GDS and obtained results analogous to those from HeLa cells, indicating that AuNP GDS-mediated delivery of AS oligo is not specific to cell lines. Addition of saturating amounts of AuNP GDS-p53-AS conjugates did not further enhance the knockdown efficiency of p53 expression (Supplementary Fig. S2†).
To investigate the ability of AuNP GDS as a general gene delivery system, we tested an additional antisense DNA that targeted Mcl-1L (myeloid cell leukemia-1 long) gene. The gene Mcl-1L encodes an anti-apoptotic Bcl-2 family protein that was discovered as an early induction gene during leukemia cell differentiation. Antisense Mcl-1L DNA contains a sequence complementary to RNA I, followed by a 20 nucleotide-sequence complementary to an internal coding region (bases 576 to 595) of the Mcl-1L mRNA.14 The AuNP GDS-Mcl-1L-AS conjugates were incubated with HeLa cells or 293T cells. After a 24-hour incubation, cells were harvested and the expressions of target proteins were quantified using Western blot analysis. The nanoconjugates efficiently decreased the levels of target proteins in both human cell lines (Supplementary Fig. S3†).
Next, we tested steady state levels of target mRNA in 293T cells treated with AuNP GDS-AS. They were decreased to less than 40% of those of untreated cells as measured using semi-quantitative RT PCR (Supplementary Fig. S4†). Based on these results, we speculate that AuNP GDS-AS mediated knockdown of gene expression stems from inhibition of translation by formation of a mRNA-antisense DNA duplex, which consequently causes a rapid degradation of target mRNAs because mRNAs are no longer protected by ribosomes from nuclease attack. In any case, these data demonstrate that AuNP GDS can be used as a general gene delivery system for antisense DNAs specific to any gene of interest in human cell lines.
We also tested another AuNP GDS that contained an oligo bearing a sequence complementary to the coding region (bases 1198 to 1215) of EGFP (an anti-EGFP oligo); the anti-EGFP oligo has been shown to be effective in the delivery and silencing of EGFP expression in human tissue cultures without interfering with expression of any other genes.8f An antisense DNA similar to AS-p53 except having a sequence complementary to the sequence of anti-EGFP oligo was annealed to this AuNP GDS and the resulting conjuate was incubated with 293T cells. Analogous results to those shown in Fig. 3 were obtained (Supplementary Fig. S5†).
To be effective as an antisense nucleic acid, the activity needs to be stably maintained for a prolonged incubation time in the cell. For this reason, we examined the incubation time-dependent efficiency of AuNP GDS-AS by measuring the degree of knockdown levels of target protein expression in HeLa and 293T cells over a three day period. The knockdown efficiency was decreased slightly from 84.5 to 78.0% for MCL-1L in 293T cells, while it was maintained between 85.3 and 94.8% for p53 in these cells after a three day incubation (Supplementary Fig. S6†). These results show that most of the activity of antisense DNAs delivered by AuNP GDS was maintained for three days in human cells.
Single or double stranded DNA oligonucleotides bound to gold nanoparticles show greatly enhanced stability relative to unbound DNA or RNA oligonucleotides in the cell.15 This stability may be due to steric hindrance and high local concentrations of salts, which can deactivate the enzymatic activity of nucleases. To test whether the activity of AS DNA oligos delivered by AuNP GDS as a duplex form is maintained longer than that of AS oligos delivered by liposome as a naked form, we measured the knockdown efficiency of AuNP GDS-AS-p53 in K562 over a five day period. K562 cells were used because they do not require adherence to culture dishes to grow, which sometimes prevented accurate measurement of the knockdown efficiency by AuNP GDS-AS. As shown in Fig. 4, the AS-p53 delivered by AuNP GDS remained active for three to four days, whereas its activity lasted for less than three days when delivered by liposome formation. The activity of AS-p53 delivered by either method was similar for two days (data not shown). These results show that AS oligos bound to gold nanoparticles are more stable than unbound AS oligos in human cells.
 | ||
| Fig. 4 Duration of knockdown effects on p53 gene expression by AS-p53 delivered by AuNP GDS and liposome. | ||
In conclusion, we have shown that single-stranded DNA functionalized gold nanoparticles (AuNP GDS) can be used as gene delivery systems for oligo antisense DNAs specific to any gene of interest without affecting normal cell physiology. The AuNP GDS-antisense conjugates efficiently knock down the expression of target proteins and, moreover, the antisense DNAs delivered by the AuNP GDS remained active for a longer period of time than those delivered by liposome formation. In addition, this system may be easily applicable to the delivery of siRNA, ribozyme, DNAzyme, and peptide nucleotide acids.
This research was supported by the Pioneer Research Program (Grant No. M1071118001-08M1118-00110) and the 21C Frontier Microbial Genomics and Application Center Program to K. Lee, Priority Research Centers (2009-0093817), National Research Foundation (2009-0071807), and the Korea Foundation for International Cooperation of Science & Technology(KICOS) 2009 (2008-00656) to M. S. Han.
Notes and references
- (a) F. McCormick, Nat. Rev. Cancer, 2001, 1, 130 CrossRef CAS; (b) J. B. Opalinska and A. M. Gewirtz, Nat. Rev. Drug Discovery, 2002, 1, 503 CrossRef CAS.
- M. A. Kay, J. C. Glorioso and L. Naldini, Nat. Med., 2001, 7, 33 CrossRef CAS.
- (a) S. E. Raper, N. Chirmule, F. S. Lee, N. A. Wivel, A. Bagg, G.-P. Gao, J. M. Wilson and M. L. Batshaw, Mol. Genet. Metab., 2003, 80, 148 CrossRef CAS; (b) S. Hacein-Bey-Abina, C. von Kalle, M. Schmidt, F. Le Deist, N. Wulffraat, E. McIntyre, I. Radford, J. L. Villeval, C. C. Fraser, M. Cavazzana-Calvo and A. Fischer, N. Engl. J. Med., 2003, 348, 255 CrossRef; (c) J. J. Green, R. Langer and D. G. Anderson, Acc. Chem. Res., 2008, 41, 749 CrossRef CAS.
- I. M. Verma and N. Somia, Nature, 1997, 389, 239 CrossRef CAS.
- (a) M. A. Mintzer and E. E. Simanek, Chem. Rev., 2009, 109, 259 CrossRef CAS; (b) X. Guo and F. C. Szoka Jr., Acc. Chem. Res., 2003, 36, 335 CrossRef CAS; (c) R. K. Tekade, P. V. Kumar and N. K. Jain, Chem. Rev., 2009, 109, 49 CrossRef CAS; (d) K. S. Soppimath, T. M. Aminabhavi, A. R. Kulkarni and W. E. Rudzinski, J. Controlled Release, 2001, 70, 1 CrossRef CAS.
- (a) M. Prato, K. Kostarelos and A. Bianco, Acc. Chem. Res., 2008, 41, 60 CrossRef CAS; (b) J. Cheon and J.-H. Lee, Acc. Chem. Res., 2008, 41, 1630 CrossRef CAS; (c) B. G. Trewyn, I. I. Slowing, S. Giri, H.-T. Chen and V. S.-Y. Lin, Acc. Chem. Res., 2007, 40, 846 CrossRef CAS; (d) I. I. Slowing, J. L. Vivero-Escoto, C.-W. Wu and V. S.-Y. Lin, Adv. Drug Delivery Rev., 2008, 60, 1278 CrossRef CAS; (e) C. J. Murphy, A. M. Gole, J. W. Stone, P. N. Sisco, A. M. Alkilany, E. C. Goldsmith and S. C. Baxter, Acc. Chem. Res., 2008, 41, 1721 CrossRef CAS.
- M.-C. Daniel and D. Astruc, Chem. Rev., 2004, 104, 293 CrossRef CAS.
- (a) K. K. Sandhu, C. M. Mclntosh, J. M. Simard, S. W. Smith and V. M. Rotello, Bioconjugate Chem., 2002, 13, 3 CrossRef CAS; (b) P. S. Ghosh, C.-K. Kim, G. Han, N. S. Forbes and V. M. Rotello, ACS Nano, 2008, 2, 2213 CrossRef CAS; (c) M. Thomas and A. M. Klibanov, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 9138 CrossRef CAS; (d) C. Agbasi-Porter, J. Ryman-Rasmussen, S. Franzen and D. Feldheim, Bioconjugate Chem., 2006, 17, 1178 CrossRef; (e) C.-Y. Tsai, A.-L. Shiau, P.-C. Cheng, D.-B. Shieh, D.-H. Chen, C.-H. Chou, C.-S. Yeh and C.-L. Wu, Nano Lett., 2004, 4, 1209 CrossRef CAS; (f) N. L. Rosi, D. A. Giljohann, C. S. Thaxton, A. K. R. Lytton-Jean, M. S. Han and C. A. Mirkin, Science, 2006, 312, 1027 CrossRef CAS.
- B. Polisky, Cell, 1988, 55, 929 CrossRef CAS.
- S. J. Hurst, A. K. R. Lytton-Jean and C. A. Mirkin, Anal. Chem., 2006, 78, 8313 CrossRef CAS.
- R. Agami and R. Bernards, Cell, 2000, 102, 55 CrossRef CAS.
- L.-C. Dai, X. Wang, X. Yao, L.-S. Min, F.-C. Qian and J.-F. He, Acta Pharmacol. Sin., 2006, 27, 1453 Search PubMed.
- (a) T. Cedervall, I. Lynch, M. Foy, T. Berggård, S. C. Donnelly, G. Cagney, S. Linse and K. A. Dawson, Angew. Chem., Int. Ed., 2007, 46, 5754 CrossRef CAS; (b) D. A. Giljohann, D. S. Seferos, P. C. Patel, J. E. Millstone, N. L. Rosi and C. A. Mirkin, Nano Lett., 2007, 7, 3818 CrossRef CAS.
- C. Skoda, B. M. Erovic, V. Wachek, L. Vormittag, F. Wrba, H. Martinek, G. Heiduschka, P. Kloimstein, E. Selzer and D. Thurnher, Oncol. Rep., 2008, 19, 1499 CAS.
- (a) Z. Wang, A. G. Kanaras, A. D. Bates, R. Cosstick and M. Brust, J. Mater. Chem., 2004, 14, 578 RSC; (b) D. S. Seferos, A. E. Prigodich, D. A. Giljohann, P. C. Patel and C. A. Mirkin, Nano Lett., 2009, 9, 308 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental details of preparation of AuNP GDS-AD conjugates, Western blot analysis, and semi-quantitative RT PCR. See DOI: 10.1039/c0cc00103a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2010 |