Quantitative, reagentless, single-step electrochemical detection of anti-DNA antibodies directly in blood serum†
Francesco
Ricci
*a,
Gianluca
Adornetto
a,
Danila
Moscone
a,
Kevin W.
Plaxco
b and
Giuseppe
Palleschi
a
aUniversity of Rome Tor Vergata, Dipartimento di Scienze e Tecnologie Chimiche, Via della Ricerca Scientifica, 00133, Rome, Italy
bDepartment of Chemistry and Biochemistry, University of California, Santa Barbara, California 93106, USA
First published on 8th January 2010
Abstract
Here we demonstrate the use of redox labeled double- and single-stranded oligonucleotides as recognition probes for the reagentless, single-step, electrochemical detection of anti-DNA antibodies directly in blood serum.
Anti-DNA antibodies are important markers for the diagnosis of several autoimmune diseases.1,2 For example, systemic lupus erythematosus (SLE), a chronic autoimmune connective tissue disease, is characterized by the production of an array of IgM and IgG autoantibodies directed against nuclear components, the most frequent targets of which are double-stranded (ds) and single-stranded (ss) DNA.3–5 Indeed, both anti-ssDNA and anti-dsDNA antibodies are involved in disease development,6 and high levels of anti-DNA antibodies are associated with disease flares (acute exacerbation of the disease).7,8 Consequently, the quantitative monitoring of sera levels of anti-DNA antibodies provides key insights into the activity and progression of the disease.9,10
Historically, several methods have been employed to quantify anti-DNA antibodies in the clinic, including the Farr assay, the Crithidia luciliae immunofluorescence technique (CLIFT), and a variety of other immunochemical approaches such as the enzyme-linked immunosorbent assay (ELISA), the bead-based immunoassay (luminex) and the fluorescent enzyme immunoassay (EliA).9,11–15 However, whereas these methods are sensitive and specific, they are also slow, cumbersome, and laboratory-bound approaches that require hours or days to return an answer to the clinician. In short, the need for a rapid, quantitative point-of-care approach to the detection of anti-DNA antibodies remains unmet.
In an effort to develop quantitative molecular diagnostics suitable for point-of-care, we have developed a reagentless, single-step electrochemical sensor platform comprised of an electrode-bound, redox-tagged oligonucleotide probe.16–19 Signalling in these electrochemical DNA (E-DNA) sensors requires only that target binding to the oligonucleotide probe changes the efficiency with which the attached redox tag strikes the electrode.20,21 Because of this, the E-DNA platform appears readily amenable to the detection of any target that binds to a DNA probe and, in doing so, changes the probe’s collisional dynamics.22 Thus inspired, we demonstrate here the use of an E-DNA-like sensor to detect anti-DNA antibodies at low nanomolar concentrations.
E-DNA sensors readily support the detection of anti-DNA antibodies. To show this, we have fabricated sensors from a 27-base probe modified with a 5′ thiol group and a methylene blue redox tag at its 3′ end. Following immobilization on a gold screen printed electrode, which supports strong chemisorption with the terminal thiol group, the probe was hybridized to a 22-base complementary sequence to form a double-stranded element (Fig. 1, left). In the absence of a target this double-stranded probe produces a significant current at the potential expected for the reduction of the methylene blue redox tag (Fig. 1, right). In the presence of anti-dsDNA antibodies this current is suppressed significantly, presumably because antibody binding reduces the flexibility of the probe and thus reduces the efficiency with which the redox tag collides with, and thus exchanges electrons with, the electrode. Upon titration with anti-dsDNA antibodies the dose-response curve of the sensor exhibits an EC50 (the analyte concentration inducing 50% of the maximum signal suppression) of 19 nM (an affinity comparable to that reported in literature23), and a detection limit of ∼10 nM (∼1.5 μg ml−1) with a linear response up to 80 nM (Fig. 2, left). A brief wash in 8 M urea is sufficient to regenerate 99% of the sensor’s original signal (Fig. SI2†), demonstrating that the observed signal decrease is not due to degradation of the sensor and allowing for ready re-use.
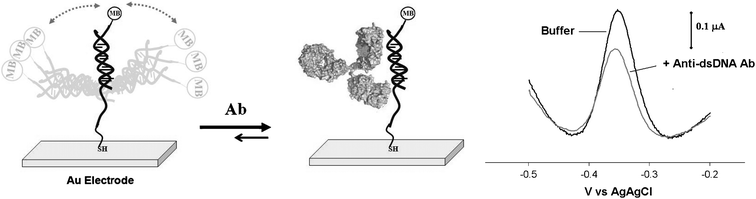 | ||
| Fig. 1 We have fabricated reagentless, single-step, E-DNA-like sensors for the detection of anti-DNA antibodies. Our approach employs a 27-base DNA probe covalently attached to a screen printed gold electrode using thiol-gold self assembled monolayer chemistry and containing a methylene blue redox tag at the 3′ end. For the detection of anti-dsDNA antibodies this probe is hybridized with its complementary target to produce a double-stranded DNA as recognition element. (left) In the absence of a target, relatively efficient collision between the label and the electrode produces a large faradaic current from the attached methylene blue. (right) The binding of the antibody to the recognition probe reduces the efficiency with which the redox reporter approaches the electrode, significantly reducing this faradaic current. The sensors readily detect anti-DNA antibodies at nanomolar concentrations (here shown the representative voltammograms obtained with an anti-dsDNA antibody concentration of 20 nM). | ||
 | ||
| Fig. 2 The E-DNA sensor readily detects both anti-dsDNA (left) and anti-ssDNA (right) antibodies at low nanomolar concentrations. Because anti-dsDNA antibodies cross-react with single-stranded DNA it is also possible to detect them using a single-stranded 27-base DNA probe alone with high sensitivity (right). The single-stranded probe’s affinity for authentic anti-ssDNA antibodies, however, is higher still (right). Both targets produce rather less signal suppression than that obtained with the double-stranded counterpart, presumably because antibody binding to the single-stranded probe does not efficiently inhibit collisions between the attached methylene blue redox tag and the electrode. The data points represent the average of replicate measurements conducted using three independently fabricated electrodes. | ||
Sensors employing single-stranded DNA probes support the detection of antibodies that bind to single-stranded DNA. For example, because anti-dsDNA antibodies recognize the deoxyribose phosphate backbone, they are known to cross-react with single-stranded DNA.24–26 Sensors employing our 27-base, single-stranded probe in the absence of its complementary strand thus also support the detection of anti-dsDNA antibodies (Fig. 2, right). And although the affinity with which the antibody binds single-stranded DNA in solution is reportedly slightly poorer than its affinity for double-stranded DNA,23,27 we observe an EC50 of just 7 nM with this probe. This poorer affinity for double-stranded DNA may arise due to the greater charge densities and steric hindrance associated with this probe. Similar signal suppression is observed when antibodies specific to single-stranded DNA (anti-ssDNA antibodies) are detected using the same 27-base, single-stranded probe, producing an EC50 of just 1.5 nM (Fig. 2, right). With both anti-dsDNA and anti-ssDNA antibodies, however, the signal suppression observed with this single-stranded probe is lower than that obtained when our double-stranded probe is challenged with anti-dsDNA antibodies, presumably because the antibody-ssDNA complex is flexible enough to produce a significant electrochemical signal.
Our sensors are specific and selective. For example, we do not detect any significant signal change after incubation of our sensors with high concentrations of either non-specific antibodies or BSA (Fig. 3, left). Indeed, our sensors are sufficiently effective in rejecting false positives arising due to the non-specific adsorption of interferents that they can be employed directly in complex samples: for example, both single-strand and double-strand sensors support the detection of anti-dsDNA and anti-ssDNA antibodies directly in antibody-spiked fetal calf serum diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 with buffered saline (Fig. 3, right).
10 with buffered saline (Fig. 3, right).
 | ||
Fig. 3 Our sensors are sufficiently specific to discriminate between the target antibody and non-specific proteins. For example, no significant signal is observed in the presence of a large excess of bovine serum albumin (BSA) (2 μM) or of a non-specific antibody (i.e. Anti-aflatoxin Ab, 1 μM) (left). Also, the sensors readily detect anti-DNA antibodies in more complex samples, such as fetal calf serum (diluted here 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 with buffered saline) at nanomolar concentrations (here shown the representative voltammograms obtained with an anti-dsDNA antibody concentration of 20 nM) (right). 10 with buffered saline) at nanomolar concentrations (here shown the representative voltammograms obtained with an anti-dsDNA antibody concentration of 20 nM) (right). | ||
Finally, our sensors are rapid and convenient, rendering them well suited for point-of care applications. Our sensors are, for example, supported on inexpensive, screen-printed electrodes and require only a simple, hand-held potentiostat for data collection. They are also rapid, exhibiting an equilibration time constant of ∼3 min (Fig. SI1†) and require neither wash steps nor the addition of exogenous reagents. These attributes compare quite favourably to those of existing methods for detecting anti-DNA antibodies.
To illustrate this we have performed more traditional electrochemical ELISA using the recognition element applied above. This entails incubation of a single-stranded DNA modified electrode with anti-dsDNA antibodies, incubation with a secondary antibody conjugated with alkaline phosphatase and the addition of 1-naphthylphosphate to enzymatically produce the electroactive 1-naphtol (all together requiring approximately 2 h). Monitoring the signal generated by 1-naphtol, which is directly proportional to the amount of anti-dsDNA antibody bound to the DNA probe, produces a classic sigmoidal dose-response curve with an EC50 of 3.5 nM (Fig. 4). While this represents an approximately two-fold improvement over the EC50 of our E-DNA-like sensor, the two approaches nevertheless achieve quite similar detection limits. The E-DNA-like sensor, however, achieves its detection limit in far less time than the ELISA approach, and without requiring any incubation step, the addition of conjugated antibody or the injection of an enzyme substrate.
 | ||
| Fig. 4 The E-DNA sensor detects anti-dsDNA antibodies with comparable sensitivity to that of a more traditional electrochemical ELISA assay. Of note, the experimental procedures required by the electrochemical immunosensor are more intensive in terms of cost, overall analysis time (2 additional steps), and reagents (additional conjugated secondary antibody and enzyme substrate needed). | ||
Here we have demonstrated a single-step, reagentless, electrochemical method for the detection of antibodies directed against single- and double-stranded DNA. Our approach is rapid, convenient and quantitative. It is also selective enough to deploy directly in the clinically relevant sample matrix: blood serum. Given these attributes, it appears that our approach is significantly more convenient—and significantly better suited for point-of-care applications—than existing methods for the detection of this important class of diagnostic markers.
Notes and references
- L. E. Munoz, U. S. Gaipl and M. Herrmann, Autoimmun. Rev., 2008, 7(8), 594–597 CrossRef CAS.
- B. D. Stollar, Crit. Rev. Biochem. Mol. Biol., 1986, 20(1), 1–36 Search PubMed.
- M. R. Arbuckle, M. T. McClain, M. V. Rubertone, R. Hal Scofield, G. J. Dennis, J. A. James and J. B. Harley, N. Engl. J. Med., 2003, 349(16), 1526–1533 CrossRef CAS.
- M. Teodorescu, Clin. Appl. Immun. Rev., 2002, 2(2), 115–128 Search PubMed.
- M. P. Madaio and K. Yanase, Journal of Autoimmunity, 1998, 11(5), 535–538 Search PubMed.
- M. Pavlovic, R. Chen, A. M. Kats, M. F. Cavallo, S. Saccocio, P. Keating and J. X. Hartmann, Ann. N. Y. Acad. Sci., 2007, 1108, 203–217 CrossRef CAS.
- K. P. Ng, J. J. Manson, A. Rahman and D. A. Isenberg, Arthrit. Care Res., 2006, 55(6), 900–904 Search PubMed.
- L. Spatz, A. Iliev, V. Saenko, L. Jones, M. Irigoyen, A. Manheimer-Lory, B. Gaynor, C. Putterman, M. Bynoe, C. Kowal, P. Kuo, J. Newman and B. Diamond, Methods, 1997, 11(1), 70–78 CrossRef CAS.
- A. Ghirardello, D. Villalta, G. Morozzi, A. Afeltra, M. Galeazzi, R. Gerli, A. Mathieu, P. L. Meroni, P. Migliorini, A. Radice, V. Riccieri, A. Ruffatti, G. D. Sebastiani, A. Tincani and A. Doria, Ann. N. Y. Acad. Sci., 2007, 1109, 401–406 CrossRef CAS.
- R. A. Sinico, B. Bollini, E. Sabadini, L. Di Toma and A. Radice, The use of laboratory tests in diagnosis and monitoring of systemic lupus erythematosus, J. Nephrol., 2002, 15(SUPPL. 6), S20–S27 Search PubMed.
- D. Isenberg and R. Smeenk, Lupus, 2002, 11(12), 797–800 CrossRef CAS.
- K. Janyapoon, P. Jivakanont, R. Surbrsing, W. Siriprapapan, T. Tachawuttiwat and S. Korbsrisate, Pathology, 2005, 37(1), 63–68 CrossRef CAS.
- T. Neogi, D. D. Gladman, D. Ibanez and M. Urowitz, J. Rheumatol., 2006, 33(9), 1785–1788 Search PubMed.
- G. S. Makowski and M. L. Ramsby, Ann. Clin. Lab. Sci., 2003, 33(2), 142–148 Search PubMed.
- K. Conrad, A. Ittenson, D. Reinhold, R. Fischer, D. Roggenbuck, T. Büttner, H.-P. Bosselmann, J. Steinbach and W. Schößler, Ann. N. Y. Acad. Sci., 2009, 1173, 180–185 CrossRef CAS.
- W. Yang, J. Y. Gerasimov and R. Y. Lai, Chem. Commun., 2009, 2902–2904 RSC.
- F. Ricci and K. W. Plaxco, Mikrochim. Acta, 2008, 163(3–4), 149–155 CrossRef CAS.
- A. Lubin and K. W. Plaxco, Acc. Chem. Res., 2009 Search PubMed , submitted.
- C. Fan, K. W. Plaxco and A. J. Heeger, Proc. Natl. Acad. Sci. U. S. A., 2003, 100(16), 9134–9137 CrossRef CAS.
- F. Ricci, R. Y. Lai, A. J. Heeger, K. W. Plaxco and J. J. Sumner, Langmuir, 2007, 23(12), 6827–6834 CrossRef CAS.
- F. Ricci, R. Y. Lai and K. W. Plaxco, Chem. Commun., 2007,(36), 3768–3770 RSC.
- F. Ricci, A. J. Bonham, A. C. Mason, N. O. Reich and K. W. Plaxco, Anal. Chem., 2009, 81, 1608–1614 CrossRef CAS.
- A. Buhl, S. Page, N. H. H. Heegaard, P. von Landenberg and P. B. Luppa, Biosens. Bioelectron., 2009, 25(1), 198–203 CrossRef CAS.
- B. H. Hahn, N. Engl. J. Med., 1998, 338(19), 1359–1368 CrossRef CAS.
- M. Herrmann, T. H. Winkler, H. Fehr and J. R. Kalden, Eur. J. Immunol., 1995, 25(7), 1897–1904 CrossRef CAS.
- J. K. Kalsi, A. C. R. Martin, Y. Hirabayashi, M. Ehrenstein, C. M. Longhurst, C. Ravirajan, M. Zvelebil, B. D. Stollar, J. M. Thornton and D. A. Isenberg, Mol. Immunol., 1996, 33(4–5), 471–483 CrossRef CAS.
- N. H. Heegaard, D. T. Olsen and K. L. Larsen, J. Chromatogr., A, 1996, 744, 285–94 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures, regeneration, time course experiment. See DOI: 10.1039/b922595a |
| This journal is © The Royal Society of Chemistry 2010 |
