Single-step covalent immobilization of oligonucleotides onto solid surface
Dileep Kumar
Kannoujia
ab,
Shakir
Ali
b and
Pradip
Nahar
*a
aInstitute of Genomics and Integrative Biology (CSIR), Mall Road, Delhi, 110 007, India. E-mail: pnahar@igib.res.in; Fax: +91-11-27667471; Tel: +91-11-27667439
bDepartment of Biochemistry, Faculty of Science, Jamia Hamdard (Hamdard University), Hamdard Nagar, New Delhi, 110 062, India
First published on 1st February 2010
Abstract
In this communication, we report a simple, single-step method for covalent immobilization of oligonucleotide probes onto an activated polystyrene surface. Polystyrene surface is activated by a photolinker, 1-fluoro-2-nitro-4-azidobenzene (FNAB) in a photochemical reaction by UV light. Amine-modified oligonucleotide probe is covalently immobilized to the activated surface by displacing the labile fluoro group of the activated polystyrene surface by the primary amino group of the aminated oligonucleotide probe. Biotinylated complementary oligonucleotide target is hybridized with the immobilized probe. The hybridized target is detected by binding streptavidin-peroxidase followed by assaying the enzyme for colorimetric signal measurement. The covalently immobilized oligonucleotide probe shows high selectivity in subsequent hybridization processes with the complementary target and clearly discriminates with single-base mismatched oligonucleotide targets. The method can immobilize amine- or thiol-modified oligonucleotide onto FNAB–activated polymer surface. The method could be potentially useful for designing DNA based biosensors or biochips and also in microtiter plate based assay systems in a cost effective manner.
Introduction
Immobilization of DNA or oligonucleotides on solid surface has become significantly important due to its applications in diagnosis of infectious or genetic diseases, gene expression analysis, detection of single nucleotide polymorphisms and mutations, DNA sequencing, forensic science, toxicology, pharmacogenomics, drug discovery, biosensing and environmental monitoring.1–5 A variety of DNA microarray devices and DNA chips have been developed that allow the detection of DNA or RNA by solid phase hybridization. These biochips are commonly based on the immobilization of DNA on a solid surface and are capable of capturing and binding the complementary DNA of interest from the small amount of sample.Commonly used methods for immobilizing a DNA or oligonucleotide to solid surfaces are adsorption, affinity interaction and covalent attachment. However, immobilization of oligonucleotides through adsorption or affinity interaction is not desirable due to its weak binding to solid surface resulting in detachment under stringent conditions and high nonspecific binding. This makes covalent methods a useful technique for immobilizing oligonucleotide to the solid surface. Over time, different strategies were evolved for engineering solid surfaces for covalent immobilization of oligonucleotides, such as gold,6 silicon,7 carbon,8,9 glass10,11 and other polymers including plastic surfaces.12–17 Different linker molecules have been used to functionalize the solid surfaces for covalent immobilization of modified oligonucleotide probes.10,17 Among them, heterobifunctional linkers are preferred for immobilization of biomolecules to avoid possible nonspecific cross-linking, which commonly occurs with homobifunctional linkers. Chrisey et al. (1996) reported the use of heterobifunctional linkers, containing modified N-hydroxysuccinimide (NHS) and maleimide functional group for covalent attachment of oligonucleotides.18 Another heterobifunctional linker, sulfosuccinimidyl 4-(N-maleimidomethyl)cyclohexane-1-carboxylate (SSMCC) was used to covalently attach amine- and thiol-modified oligonucleotides.19,20 Recently, photolinker molecules have become popular for covalent immobilization of protein21–23 and oligonucleotide sequence/DNA24 onto different solid matrices. Koch et al. (2000) have incorporated anthraquinone based photolinker during oligonucleotide synthesis; covalent immobilization to polymer support was effected by a photochemical reaction.25 In another approach, photoactivable biotin having an aryl azide group as the photo-reactive group has been used for covalent DNA immobilization on solid supports through biotin–avidin–biotin chemistry.26 However, preparation of photoactivable oligonucleotide or biotin is a time consuming, multi-step and tedious procedure.
Recently, azido bearing photolinker, 1-fluoro-2-nitro-4-azidobenzene (FNAB) has been used for rapid immobilization of proteins.21–23 However, so far this simple photolinker has not been exploited for immobilization of oligonucleotide or DNA. In this communication, we report, a single-step covalent attachment of aminated-oligonucleotide directly onto the FNAB–activated plastic surface without any additional reagents or chemicals. As any inert polymer surface can be activated by this method, oligonucleotide immobilization can be effected onto a variety of surfaces depending on the need; the only prerequisite is that the surface must have an C–H bond.
Experimental
Materials and reagents
Synthetic oligonucleotides were purchased from Midland Certified Reagent Company, Texas, USA and are shown in Table 1. Bovine serum albumin (BSA), 4-fluoro-3-nitroaniline, streptavidin-peroxidase, sodium dodecyl sulfate (SDS) and o-phenylenediamine dihydrochloride (OPD) were purchased from Sigma-Aldrich, St. Louis, MO, USA. 96-Well polystyrene microtiter plates were purchased from Greiner Bio-One, Frickenhausen, Germany. All other chemicals were of analytical grade and were purchased either from Qualigens, India or SRL, India. Reaction buffer was prepared by dissolving 1 M NaCl to phosphate buffer (0.1 M, pH 7.0 ± 0.2) and was used for diluting the reagents, immobilization and hybridization reaction. Washing buffer was prepared by adding 0.1% SDS to reaction buffer.| Oligonucleotide | Length (base) | Oligonucleotide sequence |
|---|---|---|
| Probe-0 | 20 | 5′–Biotin–ACA AGA CGT TTT ACA GTT GC-3′–NH2 |
| Probe-00 | 20 | 5′–Biotin–ACA AGA CGT TTT ACA GTT GC-3′ |
| Probe-1 | 20 | 5′–NH2–ACA AGA CGT TTT ACA GTT GC–3′ |
| Probe-2 | 20 | 5′–NH2–ATG TGG AAA ATC TCT AGC AG–3′ |
| Probe-3 | 20 | 5′–NH2–TCG GGG TTT TGG GTC TGA CG–3′ |
| Target-1 | 20 | 5′–Biotin–GCA ACT GTA AAA CGT CTT GT–3′ |
| Target-2 | 20 | 5′–Biotin–CTG CTA GAG ATT TTC CAC AT–3′ |
| Target-3 | 20 | 5′–Biotin–CGT CAG ACC CAA AAC CCC GA–3′ |
| Random | 20 | 5′–Biotin–AGT TCG ATC ATT CAT CTA AG–3′ |
| Mismatch-1 | 20 | 5′–Biotin–GCA ACT GTA CAA CGT CTT GT–3′ |
| Mismatch-2 | 20 | 5′–Biotin–GCA GCT GTA CAA CGA CTT GT–3′ |
Activation of polystyrene surface
Activation of polystyrene microtiter plates were carried out by 1-fluoro-2-nitro-4-azidobenzene (FNAB) as reported earlier.21,27 Briefly, FNAB solution (10 μM/50 μl of methanol/well) was poured to each well of a polystyrene microtiter plate and allowed to air dry in the dark. After complete evaporation of solvent, the photolinker-coated plate was exposed to UV light of 365 nm for 12 min in an UV Stratalinker (Model-2400; Stratagene, USA). After irradiating for stipulated time, the wells of the microtiter plates were washed three times with ethanol and once with methanol.Immobilization of aminated oligonucleotide
A twenty-base synthetic oligonucleotide (Probe-0: 5′–Biotin–ACA AGA CGT TTT ACA GTT GC–NH2–3′) was used to optimize the time and temperature for immobilization on polystyrene surface. Optimum temperature and time required for immobilization of aminated oligonucleotide was determined by incubating 100 μl/well of 4 nM Probe-0, separately at 30, 40, 50, 60 and 70 °C for 15, 30, 45, 60, 90 and 120 min, respectively, into the activated and untreated wells of polystyrene microtiter plates. A concentration dependent experiment was carried out by using various amounts (0.125, 0.25, 0.5, 1, 2, 4, 8 and 16 nM) of Probe-0 onto activated surface. After incubating for stipulated time the plates were washed vigorously five times with washing buffer. Wells with immobilized-oligonucleotides were treated with 200 μl/well of 2% BSA solution at 40 °C for 45 min to block unbound surface.Colorimetric detection of immobilized oligonucleotides
Immobilized-oligonucleotides were detected indirectly by binding streptavidin-peroxidase conjugate with the biotin of the immobilized oligonucleotide. Streptavidin-peroxidase conjugate binding (100 μl/well of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3000 v/v) was carried out at 37 °C in 60 min. Peroxidase attached to immobilized-oligonucleotide through biotin–streptavidin interaction was assayed by adding 100 μl of substrate-dye solution (5 μl H2O2 and 5.0 mg OPD dissolved in 12 ml of 0.2 M citrate buffer, pH 5.0). After 10 min, reaction was stopped by adding 20 μl of 5% H2SO4 and absorbance was recorded in an ELISA reader (Spectra Max, Molecular Devices, USA) at 490 nm and expressed as a mean of three observations.
3000 v/v) was carried out at 37 °C in 60 min. Peroxidase attached to immobilized-oligonucleotide through biotin–streptavidin interaction was assayed by adding 100 μl of substrate-dye solution (5 μl H2O2 and 5.0 mg OPD dissolved in 12 ml of 0.2 M citrate buffer, pH 5.0). After 10 min, reaction was stopped by adding 20 μl of 5% H2SO4 and absorbance was recorded in an ELISA reader (Spectra Max, Molecular Devices, USA) at 490 nm and expressed as a mean of three observations.
Hybridization studies
Immobilization of 4 nM Probe-1 (same oligonucleotide as Probe-0 but without biotinylation) was carried out into the activated wells of a polystyrene microtiter plate at 60 °C in 60 min and blocking at 40 °C in 45 min. Hybridization of Target-1 (complementary with the immobilized probe) and random sequence (non-complementary with the immobilized probe) both labeled with biotin at 5′–end, were carried out by allowing hybridization of targets for different time periods of 15, 30, 45, 60, 90 and 120 min, respectively at 37 °C. Hybridization temperature was optimized by incubating 4 nM of each, Target-1 and random oligonucleotides, separately with the immobilized-probe at different temperatures of 30, 40, 50, 60 and 70 °C, respectively for 90 min. In another set of experiments the amount of target-oligonucleotide was optimized by hybridizing different amounts of Target-1 oligonucleotide (0.125, 0.25, 0.5, 1, 2, 4, 8 and 16 nM) with the immobilized-Probe-1 at an optimized temperature of 50 °C in 90 min. Similarly a hybridization experiment was also performed with the random target oligonucleotide. Unhybridized target oligonucleotides were removed by washing vigorously with washing buffer. Streptavidin-peroxidase conjugate binding and peroxidase assay was carried out as mentioned above.Immobilization and hybridization of different oligonucleotides onto a polystyrene microtiter plate
Different oligonucleotide probes (Probe-1, Probe-2 and Probe-3), modified with amino group at 5′–end were immobilized into the activated wells of a polystyrene microtiter plate at 60 °C in 60 min, separately. After blocking, the probe-immobilized wells were loaded with 4 nM of respective oligonucleotide targets (Target-1, Target-2 and Target-3) and incubated at 45 °C for 90 min. Random target was used as a control for non-specific hybridization. After washing, streptavidin-peroxidase conjugate binding and enzyme assay was carried out as described above.Mismatch hybridization
Probe-1 oligonucleotide was immobilized into the activated wells of a polystyrene microtiter plate at 60 °C in 60 min. Hybridization reaction was performed with different target oligonucleotides by loading 4 nM per well of Target-1 (perfect complementary), Random (non-complementary), Mismatch-1 (single-base mismatch) and Mismatch-2 (three-base mismatches) into the Probe-1-immobilized activated wells, and incubated at 45 °C for 90 min. Hybridization was checked by streptavidin-peroxidase conjugate binding followed by enzymatic assay as mentioned above.Results and discussion
Polystyrene surface was activated by a photolinker, FNAB by UV light for covalent immobilization of a ligand. Azido group of FNAB in presence of UV light forms a highly reactive nitrine which inserts into the C–H bond of polystyrene by an insertion reaction and forms activated polystyrene possessing labile fluorine group, the other functional group of photolinker (Fig. 1A). Light-induced attachment of the photolinker to the solid surface was confirmed by X-ray photoelectron spectroscopy in our earlier study,23,28 which showed the presence of nitrogen and fluorine in the ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in the activated surface. The labile fluoro group of the activated polystyrene is easily displaced by nucleophilic (such as thiol or amine) attack of the modified oligonucleotide and forms covalent linkage between activated surface and the modified oligonucleotide. Schematic representation of immobilization of aminated oligonucleotide onto activated surface and hybridization of complementary target is shown in Fig. 1B.
1 in the activated surface. The labile fluoro group of the activated polystyrene is easily displaced by nucleophilic (such as thiol or amine) attack of the modified oligonucleotide and forms covalent linkage between activated surface and the modified oligonucleotide. Schematic representation of immobilization of aminated oligonucleotide onto activated surface and hybridization of complementary target is shown in Fig. 1B.
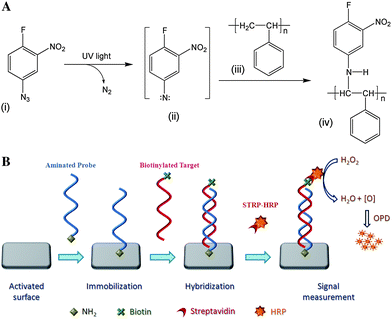 | ||
| Fig. 1 A. Schematic representation of activation of polystyrene surface by photolinker 1-fluoro-2-nitro-4-azidobenzene. B. Schematic representation of immobilization of aminated oligonucleotide probe onto an activated surface and colorimetric detection of hybridized complementary oligonucleotide target. | ||
Immobilization of oligonucleotide onto polystyrene surface was optimized by using a 20-base synthetic oligonucleotide (Probe-0) having amine modification at 3′–end and biotin at 5′–end. The amine-modified oligonucleotide was incubated at 30, 40, 50, 60 and 70 °C for 15, 30, 45, 60, 90 and 120 min, onto the activated and untreated polystyrene surface. The results shows that 60 min is the optimum time for immobilization of oligonucleotide probe onto activated polystyrene microtiter surface when the reaction is carried out at 60 °C (Fig. 2). Significant immobilization is observed in 60 min at 60 °C, beyond which no appreciable changes are observed. We noticed a sharp enhancement of immobilization kinetics beyond 40 °C. In-fact, more than a two-fold increase in absorbance is observed at 60 °C than 40 °C in 60 min, whereas, untreated surface showed insignificant immobilization (Fig. 2A). Maximum immobilization of amine-modified oligonucleotide probe onto activated surface was obtained when 8 nM of the probe is loaded. However, we have used 4 nM of the probe as it gave reasonably good qualitative assay results. Furthermore, it was necessary to overrule any non specific binding of the fluoro group of the activated surface to purine or pyrimidine base of an oligonucleotide. Thus, when biotinylated oligonucleotides probe without –NH2 modification (Probe-00) was allowed to immobilize onto activated surface, it failed to bind streptavidin-HRP conjugate to produce any assay color. On the other hand, amine modified probe (Probe-0) gave significant immobilization signal, indicating the specificity of the FNAB-activated surface towards amine modified oligonucleotides (Fig. 2B).
 | ||
| Fig. 2 A. Immobilization of aminated oligonucleotide probe carried out at different temperatures (30, 40, 50, 60 and 70 °C), each in different time periods ranging from 15 to 120 min, onto activated (closed symbols) and untreated (open symbols) wells of polystyrene microtiter plates. B. Specificity of immobilization of amine-modified oligonucleotide probe towards the activated surface. | ||
Covalent immobilization of amine-modified oligonucleotide probes onto the activated plastic surface prompted us to further investigate the efficacy of immobilization through hybridization. The results shows that hybridization of Target-1 with the immobilized Probe-1 oligonucleotide onto activated surface increases with time, whereas random target does not show any absorbance as it failed to hybridize even after a long duration (Fig. 3A). The hybridization experiment performed at 37 °C in 90 min showed significant hybridization, beyond which no appreciable increase occurred. Hybridization experiment performed at various temperatures for 90 min shows best hybridization at a temperature ranging from 40 to 50 °C. Concentration-dependent hybridization of Target-1 with immobilized-Probe-1 oligonucleotide shows that hybridization signal sharply increases upto 4 nM concentration of target oligonucleotide beyond which no further increment in hybridization signal was observed (Fig. 3B).
 | ||
| Fig. 3 A. Hybridization of perfect complementary (■) and non-complementary (▲) biotinylated target oligonucleotide with the oligonucleotide probe, immobilized onto activated wells of a polystyrene microtiter plate. B. Concentration-dependent (0.125, 0.25, 0.5, 1, 2, 4, 8 and 16 nM) hybridization of perfect complementary (■) and non-complementary (▲) targets with the probe oligonucleotide, immobilized onto activated wells of a polystyrene microtiter plate. | ||
The method was further validated by immobilizing three different oligonucleotide probes (Probe-1, Probe-2 and Probe-3) into activated wells of a polystyrene microtiter plate and subsequently hybridizing with their respective complementary oligonucleotide targets (Target-1, Target-2 and Target-3). As is shown in Fig. 4, all the target oligonucleotides perfectly hybridize with their respective complementary oligonucleotide probes. A control experiment, performed with random target does not show any hybridization signal, indicating absence of any non-specific hybridization with the immobilized-probes. Discrimination of single-base mismatched oligonucleotide from full complementary DNA is important specially in cases of disease diagnosis. Therefore specificity of the hybridization of target oligonucleotides with the immobilized-probe is further evaluated by studying hybridization of Target-1 (perfect complementary), Mismatch-1 (single-base mismatch), Mismatch-2 (three-base mismatch) and Random (non-complementary) oligonucleotide targets with immobilized-Probe-1 oligonucleotide. As shown in Fig. 5, maximum hybridization signal is observed for perfect complementary target; a significant decrease in the signal is observed in the case of single-base mismatch target and further decrease in hybridization is observed with three-base mismatch target oligonucleotide. In the case of random target no hybridization signal is noticed. Thus, the perfect complementary and single-base mismatch oligonucleotide can be easily differentiated by the present method.
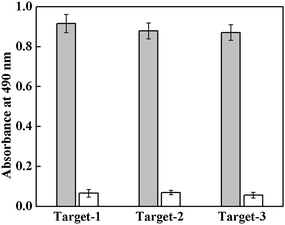 | ||
| Fig. 4 Hybridization of target oligonucleotides (Target-1, Target-2 and Target-3) with their respective complementary (Probe-1, Probe-2 and Probe-3) immobilized-probe. A random target oligonucleotide sequence was used as control for non-specific hybridization. | ||
 | ||
| Fig. 5 Hybridization of complementary (PM), single-base mismatch (MM-1), three-base mismatch (MM-2) and non-complementary (Random) target oligonucleotides with the oligonucleotide probe, immobilized onto activated wells of a polystyrene microtiter plate. | ||
Conclusions
The cost for fabrication of chip-based biosensors or DNA microarrays is one of the major limiting factors for DNA based detection systems. Herein, we have demonstrated a simple single-step method for covalent attachment of an oligonucleotide sequence onto a FNAB-activated polystyrene surface. Covalently immobilized DNA shows high selectivity in subsequent hybridization processes with the complementary target and clearly discriminates with mismatched oligonucleotide targets. The reported method for covalent immobilization of amine-modified oligonucleotide and its hybridization is a qualitative assay. Nevertheless quantitative measurement can be achieved by incorporating a fluorescent tag; thereby enhancing the applicability of this method in diagnostics and microarray. The method is versatile as it can immobilize aminated or thiol-modified oligonucleotide sequence onto any FNAB-activated polymer surface, organic or inorganic.Acknowledgements
DKK thanks the University Grant Commission, India for the award of research fellowship. The authors thank Director, Institute of Genomics and Integrative Biology (CSIR), Delhi, for his support during the course of the investigation.References
- R. W. Wallace, Mol. Med. Today, 1997, 3, 384–389 CrossRef CAS.
- G. Ramsay, Nat. Biotechnol., 1998, 16, 40–44 CrossRef CAS.
- D. J. Lockhart and E. A. Winzeler, Nature, 2000, 405, 827–836 CrossRef CAS.
- M. Schena, D. Shalon, R. W. Davis and P. O. Brown, Science, 1995, 270, 467–470 CAS.
- I. Palchetti and M. Mascini, Analyst, 2008, 133, 846–854 RSC.
- D. Peelen and L. M. Smith, Langmuir, 2005, 21, 266–271 CrossRef CAS.
- L. Zhang, T. Strother, W. Cai, X. Zhao, L. M. Smith and R. J. Hamers, Langmuir, 2002, 18, 788–796 CrossRef CAS.
- H. Cai, X. Cao, Y. Jiang, P. He and Y. Fang, Anal. Bioanal. Chem., 2003, 375, 287–293 CAS.
- M. R. Lockett, M. R. Shortreed and L. M. Smith, Langmuir, 2008, 24, 9198–9203 CrossRef CAS.
- B. Joos, H. Kuster and R. Cone, Anal. Biochem., 1997, 247, 96–101 CrossRef CAS.
- H. Sheng and B. C. Ye, Appl. Biochem. Biotechnol., 2009, 152, 54–65 CrossRef CAS.
- S. S. Ghosh and G. F. Musso, Nucleic Acids Res., 1987, 15, 5353–5372 CAS.
- S. R. Rasmussen, M. R. Larsen and S. Rasmussen, Anal. Biochem., 1991, 198, 138–142 CrossRef CAS.
- D. Liu, R. K. Perdue, L. Sun and R. M. Crooks, Langmuir, 2004, 20, 5905–5910 CrossRef CAS.
- Y. Li, Z. Wang, L. M. Ou and H. Z. Yu, Anal. Chem., 2007, 79, 426–433 CrossRef CAS.
- S. L. Beaucage, Curr. Med. Chem., 2001, 8, 1213–1244 CAS.
- F. Fixe, M. Dufva, P. Telleman and C. B. V. Christensen, Nucleic Acids Res., 2004, 32, e9 CrossRef CAS.
- L. Chrisey, G. Lee and C. O'Ferrall, Nucleic Acids Res., 1996, 24, 3031–3039 CrossRef CAS.
- J. Brockman, A. Frutos and R. Corn, J. Am. Chem. Soc., 1999, 121, 8044–8051 CrossRef CAS.
- C. Jordan, A. Frutos, A. Thiel and R. Corn, Anal. Chem., 1997, 69, 4939–4947 CrossRef CAS.
- P. Nahar, N. M. Wali and R. P. Gandhi, Anal. Biochem., 2001, 294, 148–153 CrossRef CAS.
- A. Naqvi, P. Nahar and R. P. Gandhi, Anal. Biochem., 2002, 30, 674–678.
- U. Bora, P. Sharma, S. Kumar, K. Kannan and P. Nahar, Talanta, 2006, 70, 624–629 CrossRef CAS.
- D. Dankbar and G. Gauglitz, Anal. Bioanal. Chem., 2006, 386, 1967–1974 CrossRef CAS.
- T. Koch, N. Jacobsen, J. Fensholdt, U. Boas, M. Fenger and M. Jakobsen, Bioconjugate Chem., 2000, 11, 474–483 CrossRef CAS.
- X. Su, Biochem. Biophys. Res. Commun., 2002, 290, 962–966 CrossRef.
- A. Naqvi and P. Nahar, Anal. Biochem., 2004, 327, 68–73 CrossRef CAS.
- U. Bora, K. Kannan and P. Nahar, J. Membr. Sci., 2005, 250, 215–222 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2010 |
