An ancestral luciferase in the Malpighi tubules of a non-bioluminescent beetle!
Received 1st October 2008, Accepted 20th November 2008
First published on 9th December 2008
Abstract
The evolutionary origin of beetle bioluminescence is enigmatic. Previously, weak luciferase activity was found in the non-bioluminescent larvae of Tenebrio molitor (Coleoptera: Tenebrionidae), but the detailed tissular origin and identity of the luciferase-like enzyme remained unknown. Using a closely related giant mealworm, Zophobas morio, here we show that the luciferase-like enzyme is located in the Malpighi tubules. cDNA cloning of this luciferase like enzyme, showed that it is a short AMP-ligase with weak luciferase activity which diverged long ago from beetle luciferases. The results indicate that the potential for bioluminescence in AMP-ligases is very ancient and provide a first reasonable protoluciferase model to investigate the origin and evolution of beetle luciferases.
Introduction
The evolutionary origin of bioluminescence, and of beetle bioluminescence in particular, is of evident interest.1,2 Luciferases, the enzymes that catalyze the production of bioluminescence, are a diverse group of evolutionarily unrelated oxygenases catalyzing the oxidation of distinct luciferins.3,4 In the case of bioluminescent beetles, luciferases evolved from AMP-CoA ligases,1,2 an ancient group of enzymes that catalyze (i) the activation of carboxylic substrates through adenylation at the expense of ATP, and (ii) the subsequent transfer and thioesterification of the carboxylic group to CoA (step 2). In beetle luciferases, however, the second-half reaction is replaced by the oxygenase activity, which ultimately leads to the bioluminescence function. How these luciferases evolved from AMP-ligase is not known.
| (1) X–COOH + ATP ⇌ X–COAMP + PPi + H2O (2) X–COAMP + HS–CoA ⇌ X–S–CoA + H2O + AMP |
X–COAMP + O2→ X–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O + AMP + CO2+ light O + AMP + CO2+ light |
Twelve years ago, an enzyme able to catalyze the emission of weak light in the presence of firefly D-luciferin and Mg/ATP or of luciferyl adenylate was reported in larvae of the non-luminous beetle Tenebrio molitor.5 While the sequence of this luciferase-active protein is still unknown, several genes homologous to firefly luciferases, perhaps resulting from gene duplication, were recently found in the same mealworm and subsequently cloned; however, the proteins they expressed showed acyl-coenzymeA synthetic activity, but no luciferase activity.6
We report here the anatomic location, cloning, sequence analysis and properties of the protein responsible for light emission upon injection of firefly D-luciferin in a closely related beetle, the giant mealworm Zophobas morio (Coleoptera: Tenebrionidae) (Fig. 1, see later).
Experimental
Insects
Live Zophobas morio and Tenebrio molitor mealworms were commercially available in pet shops.CCD Imaging
Mealworm weak chemiluminescence was imaged after injecting 100 μL of 10 mM D-luciferin in citrate buffer pH 5 by exposure during 5 min with a Xenogen IVIS CCD camera system (CA, USA), or by 20–30 min exposure with an ATTO Light-Capture I CCD camera system (Tokyo, Japan). Controls using only citrate buffer instead of luciferin, did not produced luminescence. The Malpighi tubules were isolated from the live larvae previously injected with D-luciferin, and drawn in 100 μL of a solution consisting of 7.1 mM NaH2PO4, 4.9 mM MgCl2, 22.3 mM MgSO4, 4 mM CaCl2, 260 mM sucrose, 8.8 mM glucose before imaging.cDNA cloning
A cDNA library was constructed into UNI-ZAP vector (Stratagene, La Jolla, CA) using mRNA isolated from the Malpighi tubules (Mini RNAm kit; QIAGEN). The amplified phage library was excised in E.coli SOLR cells to yield the phagemid library, and the transfected into SOLR cells to produce bacterial colonies with plasmids. The bacterial colonies were transferred to nitrocellulose membranes, induced with IPTG at 20 °C overnight, sprayed with 10 mM D-luciferin pH 5, and finally exposed in Xenogen IVIS CCD camera (5 min). One positive clone was isolated and sequenced using universal M13 and internal primers. Recombinant luciferase-like enzyme was expressed in E.coli BL21-DE3 cells, after IPTG induction at 20 °C overnight.The luciferase-like enzyme was extracted from the mealworm Malpighi tubules in cold extraction buffer consisting of 0.09 M sodium phosphate buffer, 10% glycerol, 1% Triton X-100, 1 mM EDTA, 1 mM DTT, pH 7.5. In the case of the recombinant enzyme, bacterial pellets expressing the enzyme were extracted in the same buffer or in 25 mM Tris-HCl buffer pH 8 containing 10% glycerol, 1% Triton X-100, 1 mM EDTA, 1 mM DTT by 3 freeze–thawing cycles in dry ice. The extracts were then centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g during 15 min at 4 °C. The supernatants, called crude extracts, were removed and used in the luminescence assays. The luminescence assays were carried out by mixing 10–20 μL of crude extracts to 80 μL of assay buffer consisting of 0.1M Tris pH 8.5, 2 mM D-luciferin, 2 mM ATP, 4 mM MgSO4 in a luminometer tube. Assays for luciferyl-adenylate chemiluminescence were performed by mixing 90 μL of Tris-HCl 0.10 M pH 8.5, 10 μL of crude extract and 10 μL of luciferyl-adenylate. Luminescence activity was measured using TDIII (Tokyo, Japan) and AB2200 (ATTO; Tokyo, Japan) luminometers.5
000 g during 15 min at 4 °C. The supernatants, called crude extracts, were removed and used in the luminescence assays. The luminescence assays were carried out by mixing 10–20 μL of crude extracts to 80 μL of assay buffer consisting of 0.1M Tris pH 8.5, 2 mM D-luciferin, 2 mM ATP, 4 mM MgSO4 in a luminometer tube. Assays for luciferyl-adenylate chemiluminescence were performed by mixing 90 μL of Tris-HCl 0.10 M pH 8.5, 10 μL of crude extract and 10 μL of luciferyl-adenylate. Luminescence activity was measured using TDIII (Tokyo, Japan) and AB2200 (ATTO; Tokyo, Japan) luminometers.5Preparation of luciferyl-adenylate
The luciferyl-adenylate from D-luciferin and AMP was prepared as previously described.7Western blots
Western blots were done for Malpighi tubules crude extracts and for bacteria expressing the recombinant luciferase-like enzyme by using polyclonal antibodies raised against firefly or Phrixtrix hirtus railroadworm luciferases using ECL kit (Amersham, Buckinghamshire, UK).Results and discussion
Anatomic location of luciferase-like enzyme
Careful dissection of Zophobas morio mealworm showed that the light emission originates from the Malpighi tubules (Fig. 1). Crude extracts of Malpighi tubules emitted light in the presence of D-luciferin and ATP/Mg, and displayed antigenic bands in Western blots using polyclonal antibodies against beetle luciferases.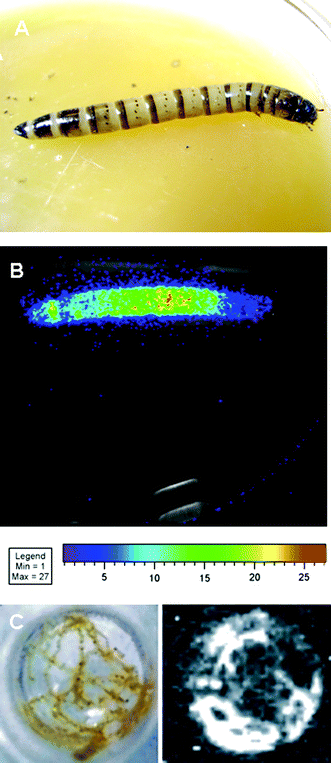 |
| | Fig. 1 CCD imaging of Zophobas morio mealworm induced chemiluminescence after injection of firefly D-luciferin: (A) live mealworm; (B) mealworm chemiluminescence image after intra-cuticular injection of 100 μL of 10 mM D-luciferin in 0.1M citrate buffer pH 5.0 (5 min exposure with a Xenogen model IVIS CCD camera system); (C) Malpighi tubules isolated from D-luciferin injected mealworms suspended in an incorporation solution (7 mM NaH2PO4, 5 mM MgCl2, 22 mM MgSO4, 4 mM CaCl2, 260 mM sucrose, 9 mM glucose); (D) chemiluminescence image of C obtained with an ATTO (Tokyo, Japan) model Light-Capture I CCD camera system after 30 min exposure at high sensitivity. | |
cDNA structure
A cDNA library was constructed from the mRNA isolated from Malpighi tubules. Colonies were tested for emission in the presence of D-luciferin using a CCD camera. A weak emitting positive clone was isolated and its cDNA sequenced. It consists of 1733 bp coding for a polypeptide of 528 residues (Fig. 2). The N-terminal and C-terminal halves have endoplasmic reticulum membrane signal-like peptides SARI (Ser-Ala-Arg-Ile) and AEKF (Ala-Glu-Lys-Phe), respectively, instead of the C-terminal SKL (Ser-Lys-Leu) peroxissomal targeting peptide found in beetle luciferases. |
| | Fig. 2 Amino-acid sequences alignment of Zophobas morio luciferase-like enzyme and related beetle luciferases. Multialignement was done using CLUSTALW2 program. (Ppy) Photinus pyralis firefly luciferase; (Lcr) Luciola cruciata firefly luciferase; (PxRE) Phrixotrix hirtus railroadworm red-emitting luciferase; (Pte) Pyrearinus termitilluminans larval click beetle luciferase; (Zop) Zophobas morio luciferase-like enzyme. (Gray shadow) ATP-binding and adenylation motifs and (yellow shadow) luciferin-binding motifs. | |
Phylogeny
This luciferease-like enzyme is 26–32% identical to that of beetle luciferases but 14–20 residues shorter. The highest sequence identities were found with similarly sized AMP-CoA ligases from the tenebrionids Tribolium casteneum (51–65%) and Tenebrio molitor mealworm (29–51%), and from mosquitoes. Noteworthy, in the phylogenetic analysis this protein appears closer to benzoate CoA ligases rather than beetle luciferases and their close homologs (Fig. 3). |
| | Fig. 3 Unrooted phylogenetic tree of Zophobas morio luciferase-like enzyme, beetle luciferases and related AMP-ligases. Multiple sequence alignments were performed using Clustal W 1.8 and default parameters and Blosum. Phylogenetic analysis was performed with PAUP* 4.0b1017 using neighbor-joining method18 with an adapted version of Blosum 35. The phylogenetic tree was visualized using T-Rex.19 | |
Luminescence properties
Table 1 summarizes some physical-chemical properties of the crude extracts recombinant enzyme. Bacterial extracts expressing the recombinant luciferase-like enzyme displayed weak luminescence in presence of either D-luciferin and MgATP or luciferyl-adenylate, with a peak at ∼600 nm. A very rough estimate of its quantum efficiency would place it 2 or 3 orders of magnitude lower than the yield of firefly luciferase. However, it is definitely orders of magnitude higher than the efficiency reported for the non-specific enhancement of the luciferyl adenylate chemiluminescence by BSA.7 Dilution of the crude extract results in a linear decrease of luminescence intensity, indicating that the luminescence is generated by the enzyme and not by the non-specific enhancement of chemiluminescence of released luciferyl-adenylate by other proteins, which would result in exponential decrease of luminescence. The KM value for luciferin is at least one order of magnitude higher than that measured for beetle luciferases, indicating a very low affinity for this substrate. The properties for Zophobas recombinant enzyme are very similar to the previously published data from Tenebrio molitor mealworm crude extracts,6 which were prepared from impure fat body containing Malpighi tubules preparations.
Table 1 Properties of Zophobas morii luciferase-like enzyme and related enzymes
| Enzyme | Aspa/V mg−1 | KM LH2b/μM | λmaxc/nm |
|---|
| Specific bioluminescence activity measured in V per mg of protein. Apparent KM for D-luciferin. Peak of bioluminescence spectrum. |
|---|
| Tenebrio molitor extracts | | 800 | 610 |
| Zophobas mori Luc | 0.02 | 260 | 600 |
| |
| Firefly luciferases |
| Photinus pyralis | 23 | 10 | 558 |
| |
| Railroad worm luciferases |
| Phrixotrix hirtus | 1.2 | 7 | 623 |
| Phrixotrix vivianii | 2.3 | 64 | 546 |
| |
| Click beetle luciferases |
| P. termitilluminans | 7.2 | 65 | 534 |
Origin of luciferases from AMP/CoA ligases
It is, of course, impossible to assert that this AMP-ligase with luciferase activity from the Malpighi tubules of Zophobas is in any way a direct ancestor of firefly luciferases. It is clear, however, that the potential for light emission was present very early during the evolution of AMP-ligases, perhaps before the divergence of the main Eukaryotic phyles. The potential for light emission, clearly appeared in the first enzymes able to catalyze the adenylation of D-luciferin or similar substrates, producing a weak red chemiluminescence that could be easily enhanced by hydrophobic protein binding sites. Thus, it is likely that during the first incipient stages of bioluminescence evolution (protobioluminescence), light emission was in the red region.As expected, this enzyme displays the signature motifs of AMP-ligases. When compared with beetle luciferases,8,9 however, this enzyme usually displays shorter loops. According to Photinus pyralis firefly luciferase amino-acid sequence numbering, the putative luciferin-binding site displays the following substitutions in Zophobas luciferase-like enzyme: R218I, F243M, F/H244Y, H245W, E311C and R331L (Fig. 2). The corresponding residues R218, F/H244 and H245 in firefly luciferases are known to be important for luciferin phenolate, benzothiazole and thiazole rings binding and stabilization, and for oxygenase activity.10,11 The loops between residues 223–235 and 354–362, which are important to keep the active site structure and for bioluminescence colors,12,13 showed a 5 residues-deletion and a 4-residues insertion, respectively. Noteworthy, previously we have shown that deletion mutagenesis of the residues G225 and T226 (G228 and N229 in P. pyralis firefly luciferase) in the loop between residues 223–235 of Pyrearinus termitilluminans click beetle luciferase, resulted in very weak red-emitting mutants. Whether all these substitutions are responsible for the low affinity for luciferin and for the weak luciferase activity in Zophobas luciferase-like enzyme remains to be investigated.Biological function of the luciferase-like enzyme
The AMP-ligase and luminescence activity of the mealworm luciferase-like enzyme in presence of D-luciferin is by no means accidental, since mealworms have not this substrate as we have extensively investigated. The AMP-ligases are known for their role in activating a wide variety of carboxylic acids for different purposes, including transport across membranes, biosynthetic pathways, degradation pathways and detoxification of xenobiotics.2 Several AMP-ligases are promiscuous in relation to their carboxylic substrates. The location of the enzyme in the Malpighi tubules, active in the excretion; its association with the endoplasmic reticulum, which can be involved in drug detoxification; and its similarity with bacterial benzoate-CoA ligases are consistent with the involvement some AMP-ligases in excretion and detoxification of carboxylic acids.14,15 Several compounds are found to be excreted in the lumen of Malpighi tubules, including pteridines and carboxylic acids.15 One of the pathways of detoxification and excretion of carboxylic acid xenobiotics is the conjugation with amino-acids via tioesterification with CoA in reactions catalyzed by xenobiotic CoA ligases, the so called Fase II reactions.14Zophobas luciferase-like enzyme could be one of these AMP-ligases involved in excretion and detoxification of a range of structurally related substrates. Remarkably, the bioluminescence of the distantly related Dipteran, Arachnocampa ssp, originates also from Malpighi tubules, and is also ATP dependent.16
Acknowledgements
The author (V. R. V.) wish to dedicate this work to Sophia K. Viviani. We deeply thank J. W. Hastings and Thérèse Wilson (Harvard University) for their continuous support, E. J. H. Bechara (São Paulo University, Brazil) and Y. Ohmiya (AIST, Osaka, Japan) for many discussions and encouragement. We are also indebted to Laboratório Nacional de Luz Síncrotron (LNLS, Campinas, SP, Brazil) for providing sequencing facilities and to Gabriela Pitondo Reis for technical assistance. This work was funded by grants from Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP, Brazil) and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brazil).References
- K. V. Wood, The chemical mechanism and evolutionary development of beetle bioluminescence, Photochem. Photobiol., 1995, 62, 662–673 CrossRef CAS.
- V. R. Viviani, The Origin, Diversity and Structure Function Relationships of Insects Luciferases, Cell. Mol. Life Sci, 2002, 59, 1833–1850 CrossRef CAS.
- J. W. Hastings, Biological diversity, chemical mechanisms and evolutionary origins of bioluminescent systems, J. Molecular Evolution, 1983, 19, 309–321 Search PubMed.
- J. F. Rees, B. De Wergifosse, O. Noiset, M. Debuisson, B. Janssens and M. E. Thompson, J. Exp. Biol, 1998, 201, 1211 Search PubMed.
- V. R. Viviani and J. H. Bechara, Larval Tenebrio molitor (Coleoptera: Tenebrionidae) fat body extracts catalyze D-luciferin and ATP-dependent chemiluminescence. A luciferase-like enzyme, Photochem. Photobiol., 1996, 63, 713–718 CrossRef CAS.
- Y. Oba, M. Sato and S. Inouye, Cloning and characterization of the homologous genes of firefly luciferase in the mealworm beetle, Tenebrio molitor, Insect Mol. Biol., 2006, 15, 293–299 CrossRef CAS.
- V. R. Viviani and Y. Ohmiya, Bovine serum albumin displays luciferase-like activity in presence of luciferyl-adenylate: insights on the origin of protoluciferase activity and bioluminescence colors, Luminescence, 2006, 21, 262–267 CrossRef CAS.
- E. Conti, N. P. Franks and P. Brick, Crystal Structure of Firefly Luciferase Throws Light on a Super Family of Adenylate-forming Enzymes, Structure, 1996, 4, 287–298 CrossRef CAS.
- T. Nakatsu, S. Ichiyama, J. Hiratake, A. Saldanha, N. Kobashi, K. Sakata and H. Kato, Structural basis for the spectral difference in luciferase bioluminescence, Nature, 2006, 440, 372–376 CrossRef CAS.
- B. R. Branchini, R. A. Magyar, M. H. Murtishaw, S. M. Anderson and M. Zimmer, Site-directed mutagenesis for histidine 245 in firefly luciferase: a proposed model of the active-site, Biochemistry, 1998, 37, 15311–15319 CrossRef CAS.
- B. R. Branchini, R. A. Magyar, M. H. Murtishaw and N. C. Portier, The role of active site residue arginine 218 in firefly bioluminescence, Biochemistry, 2001, 40, 2410–2418 CrossRef CAS.
- V. R. Viviani, A. J. Silva-Neto, F. G. C. Arnoldi, J. A. R. G. Barbosa and Y. Ohmiya, The influence of the loop between residues 223–235 in beetle luciferase bioluminescence spectra: a solvent gate for the active site of pH-sensitive luciferases, Photochem. Photobiol., 2007, 83, 1–7 Search PubMed.
- V. R. Viviani, F. M. Okawachi, V. Scorsato and F. C. Abdalla, CCD imaging of basal bioluminescence in larval fireflies: clues on the anatomic origin and evolution of bioluminescence, Photochem. Photobiol. Sci., 2008, 7, 448–452 RSC.
- K. M. Knights and C. J. Drogemuller, Xenobiotic-CoA ligases: kinetic and molecular characterization, Current Drug Metab., 2000, 1, 49–66 Search PubMed.
- W. C. Dauterman, in Comprehensive Insect Physiology, Biochemistry and Pharmacology, ed. G. A. Kerkut and L. I. Gilbert, Pergamon Press, New York, 1985, 18, pp. 713–730 Search PubMed.
- J. B. Gattenby, The Australasian mycetophilid glow-worms., Trans. R. Soc. N. Z., 1960, 88, 577–593 Search PubMed.
- D. L. Swofford, PAUP. Phylogenetic, Analysis Using Parsimony (and Other Methods),. Version 4, Sinauer Associates, Sunderland, Massachusetts, 1998 Search PubMed.
- N. Saitou and M. Nei, The neighbor-joining method: a new method for reconstruction of phylogenetic trees., Mol. Biol. Evol., 1987, 4, 406–425 Search PubMed.
- V. Makarenkov, T-Rex: reconstructing and visualizing phylogenetic trees and reticulation networks, Bioinformatics, 2001, 17, 664–668 CrossRef CAS.
|
| This journal is © The Royal Society of Chemistry and Owner Societies 2009 |
Click here to see how this site uses Cookies. View our privacy policy here. ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O + AMP + CO2+ light
O + AMP + CO2+ light![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g during 15 min at 4 °C. The supernatants, called crude extracts, were removed and used in the luminescence assays. The luminescence assays were carried out by mixing 10–20 μL of crude extracts to 80 μL of assay buffer consisting of 0.1M Tris pH 8.5, 2 mM D-luciferin, 2 mM ATP, 4 mM MgSO4 in a luminometer tube. Assays for luciferyl-adenylate chemiluminescence were performed by mixing 90 μL of Tris-HCl 0.10 M pH 8.5, 10 μL of crude extract and 10 μL of luciferyl-adenylate. Luminescence activity was measured using TDIII (Tokyo, Japan) and AB2200 (ATTO; Tokyo, Japan) luminometers.5
000 g during 15 min at 4 °C. The supernatants, called crude extracts, were removed and used in the luminescence assays. The luminescence assays were carried out by mixing 10–20 μL of crude extracts to 80 μL of assay buffer consisting of 0.1M Tris pH 8.5, 2 mM D-luciferin, 2 mM ATP, 4 mM MgSO4 in a luminometer tube. Assays for luciferyl-adenylate chemiluminescence were performed by mixing 90 μL of Tris-HCl 0.10 M pH 8.5, 10 μL of crude extract and 10 μL of luciferyl-adenylate. Luminescence activity was measured using TDIII (Tokyo, Japan) and AB2200 (ATTO; Tokyo, Japan) luminometers.5


