Analytical methods to assess nanoparticle toxicity
Bryce J.
Marquis
,
Sara A.
Love
,
Katherine L.
Braun
and
Christy L.
Haynes
*
Department of Chemistry, University of Minnesota, Minneapolis, MN 55455, USA. E-mail: chaynes@umn.edu; Web: http://www.chem.umn.edu/groups/haynes/ Fax: +1 612-626-7541; Tel: +1 612-626-1096
First published on 5th February 2009
Abstract
During the past 20 years, improvements in nanoscale materials synthesis and characterization have given scientists great control over the fabrication of materials with features between 1 and 100 nm, unlocking many unique size-dependent properties and, thus, promising many new and/or improved technologies. Recent years have found the integration of such materials into commercial goods; a current estimate suggests there are over 800 nanoparticle-containing consumer products (The Project on Emerging Nanotechnologies Consumer Products Inventory, http://www.nanotechproject.org/inventories/consumer/, accessed Oct. 2008), accounting for 147 billion USD in products in 2007 (Nanomaterials state of the market Q3 2008: stealth success, broad impact, Lux Research Inc., New York, NY, 2008). Despite this increase in the prevalence of engineered nanomaterials, there is little known about their potential impacts on environmental health and safety. The field of nanotoxicology has formed in response to this lack of information and resulted in a flurry of research studies. Nanotoxicology relies on many analytical methods for the characterization of nanomaterials as well as their impacts on in vitro and in vivo function. This review provides a critical overview of these techniques from the perspective of an analytical chemist, and is intended to be used as a reference for scientists interested in conducting nanotoxicological research as well as those interested in nanotoxicological assay development.
 Bryce Marquis | Bryce Marquis is currently a Ph.D. candidate in Analytical Chemistry at the University of Minnesota working under the direction of Christy Haynes. Previously, he earned an M.S. in Chemistry from Carnegie Mellon University in Pittsburgh, PA working with Neil Donahue, and a B.S. in Chemistry from Albion College in Albion, MI. His research interests include analytical methods for in vitro toxicological analysis and analysis of single-cells using microelectrodes. |
 Sara Love | Sara Love is currently a Ph.D. candidate in Chemistry at the University of Minnesota under the direction of Christy Haynes. Previously, she earned a B.A. in Chemistry at Hamline University in St. Paul, MN and an M.S. in Chemistry at the University of Minnesota with Christy Haynes. Her research interests included assessment of nanoparticle toxicity and immunogenicity. |
 Katherine Braun | Katherine Braun is an undergraduate research assistant under Professor Christy Haynes. She is working toward a degree in biomedical engineering at the University of Minnesota. Her research interests include biological microscopy and in vitro nanoparticle uptake and localization. |
 Christy Haynes | Christy L. Haynes is currently a McKnight Land-Grant Assistant Professor of Chemistry at the University of Minnesota in Minneapolis. She completed her Ph.D. work at Northwestern University with Richard P. Van Duyne. She was also a National Institutes of Health postdoctoral fellow with R. Mark Wightman at the University of North Carolina, Chapel Hill before beginning her independent career. The Haynes group research activities include developing electrochemical assays to monitor nanoparticle toxicity, exploring how immune cells communicate with one another at the single-cell level, and developing surface-enhanced Raman scattering chemosensors. |
1. Introduction
Nanotoxicology is an emerging discipline attempting to characterize and categorize the health effects caused by engineered nanomaterials in order to determine structure/function relationships between nanoparticles and toxicity. These relationships will be used to formulate a set of design rules for the design of safe nanomaterials. The increased prevalence of these materials in consumer goods has brought nanotoxicology to the forefront and has called attention to the gap in toxicological information regarding these materials. Although there has recently been a tremendous amount of activity in this field,1,2 there are a large number of significant challenges that must be overcome to allow the safe incorporation of nanomaterials into consumer goods as outlined in a recent commentary.3 From an analytical chemistry perspective, this emerging discipline poses many interesting challenges which will draw from both nanomaterial characterization and bioanalytical chemistry expertise within the field. The major goal of this review is to summarize the analytical techniques currently being used in nanotoxicology and to provide critical insight into this field. First, it is intended to act as a current summary of the analytical techniques used in nanotoxicology to provide scientists interested in conducting nanotoxicological research with a reference for conducting and comparing the results of future nanotoxicity experiments. Second, it is also intended to serve as a critical review of current nanotoxicology assessment techniques, focusing on the strengths and limitations of each technique for researchers interested in developing new nanotoxicological techniques. The techniques reviewed herein are categorized by their in vivo or in vitro application, although complete assessment requires complementary use of in vitro and in vivo methodology. The methods are organized into the following sections: characterization of cellular uptake of nanoparticles, in vitro toxicity characterization, and in vivo toxicity studies. Finally, the current and future challenges for analytical scientists in this field are discussed.2. In vitro uptake and localization characterization
Characterization of in vitro nanoparticle uptake and localization is intrinsically linked to cytotoxicological studies because uptake provides evidence of nanoparticle-cell interaction, wherein the delicate intracellular machinery is exposed to nanoparticles. Quantifying the amount of nanomaterial taken up by cells and describing the localization of these materials presents analytical challenges that research groups have approached with a variety of techniques. Based on limited technology, quantification of nanoparticle uptake is often obtained at the cost of spatial resolution, and intracellular localization information is often qualitative in nature. Due to these limitations, techniques are best used in concert with each other to provide a more complete understanding of uptake.2.1 Transmission electron microscopy
Transmission electron microscopy (TEM) can provide the most detailed information regarding in vitro nanoparticle uptake and localization by allowing both visualization of nanoparticle location within a cell or tissue and, in conjunction with spectroscopic methods, characterization of the composition of the internalized nanoparticles. Visualization is most easily accomplished with electron-dense nanomaterials, such as metal nanoparticles (Fig. 1). TEM images provide poor resolution of diffuse electron materials, as was found in studies of PEG-coated iron oxide nanoparticles; in this case, dynamic light scattering (DLS) measurements were performed in conjunction with TEM to determine the size of the nanoparticle including both the shell and core.4,5 High resolution transmission electron microscopy (HRTEM) can be used to identify the crystalline structures of particles, as was done for superparamagnetic iron oxide nanoparticles by Petri-Fink et al.6 and quartz particles by Warheit et al.7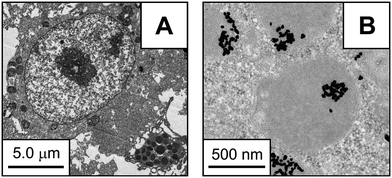 | ||
| Fig. 1 (A) TEM image of mouse peritoneal leukocyte after 48 h exposure to 1 nM 28.1 ± 6.7 nm citrate-capped gold colloids. (B) High magnification TEM image of the same leukocyte showing internalized colloidal gold. (Reprinted with permission from ref. 151. Copyright 2008, American Chemical Society.) | ||
Qualitative elemental analysis techniques can be performed on biological samples exposed to nanoparticles using an electron microscope coupled with a microanalysis system to identify the chemical composition of the nanoparticles present in the sample.8,9 For example, electron-dispersive X-ray analysis (EDS) was used to confirm the presence of silver nanoparticles within cells,10 and electron energy loss spectroscopy (EELS) was used in conjunction with TEM for elemental confirmation of carbon nanotube uptake.11 Elemental analysis is especially important because the staining procedures used to visualize biological samples may introduce electron-dense nanosized aggregates as artifacts, and it would be possible to mistake these stain artifacts for the purposefully introduced engineered nanoparticles.
Many groups use TEM to characterize nanoparticles both before and after exposure to cells7,12–18 to ensure that the morphology and size distribution of the nanoparticle does not change after exposure and uptake. However, the long time required for both biological sample preparation and image analysis greatly limits the analytical throughput when using this technique for nanoparticle uptake analysis.
2.2 Elemental analysis
Since many nanomaterials are constructed of materials not found natively in the body, uptake of these nanomaterials can be quantified by determining the mass or concentration of these non-native elements within cells. Inductively coupled plasma atomic emission spectroscopy (ICP-AES) is widely regarded as a powerful technique for the quantification of internalized nanoparticle (NP) elemental composition19,20 and has been used to quantify nanoparticle uptake. ICP-AES benefits from sub-ppb detection limits, high precision, and a dynamic range of five orders of magnitude or more.21 To date, in vitro nanotoxicity studies have used ICP-AES to assess nanoparticle uptake of gold,22–24 cerium-oxide,25 and iron-oxide nanomaterials26,27 among others. Generally, sample preparation includes isolation of cells from culture media followed by acidic sample digestion before dilution and ICP-AES analysis. In some instances, the mass concentrations obtained from ICP-AES are converted into nanoparticle numbers after estimating the mass of a single nanoparticle using the atomic weight, the crystal lattice unit length and the well-defined geometry of the nanoparticle.28 One major limitation of ICP-AES analysis in biological systems is that it is unable to characterize the uptake of carbonaceous nanoparticles (e.g. polymeric NPs and carbon nanotubes) because it cannot distinguish between cellular and nanoparticle sources of carbon. In addition, because ICP-AES is an elemental analysis method, this technique is unable to differentiate between elements confined within the nanostructure and solvated ions that had leached from the nanomaterial. Additionally, conversion of the ICP-AES measurement to standard nanoparticle dosing units requires a well-characterized crystalline lattice and nanoparticle monodispersity as well as the assumption that the nanostructure does not change upon exposure to the cellular environment. Finally, this technique does not yield any spatial information and is unable to distinguish nanoparticles associated with the exterior of the cell from those within the interior or nanoparticles associated with different cell types within a heterogeneous culture.2.3 Fluorescence spectroscopy
Fluorescence spectroscopy is useful in both the quantitative assessment of nanoparticle uptake as well as the qualitative assessment of nanoparticle localization. Quantitative assessment can be achieved in a manner similar to ICP-AES through use of bulk fluorescence29–31 or on a cell-to-cell basis using confocal fluorescence.32 It is also possible to correlate nanoparticle counting directly with cell numbers or sort cells based on nanoparticle uptake using a flow cytometry technique such as fluorescence-activated cell sorting (FACS).15,33–36 Detection is limited by the fluorescent properties of the nanoparticle species and the collection efficiency of the instrumentation while the spatial resolution is diffraction-limited, ∼200 nm at best. The inherent fluorescent properties of some nanoparticles allow for facile analysis of uptake such as the recently reported uptake of quantum dots (QDs) by stem cells.37 Recent advances in confocal light collection, such as spinning-disk confocal microscopy, can be used to monitor the trafficking of quantum dot trajectories38 within cells with sub-second time resolution.Non-natively fluorescent nanostructures can still be evaluated using fluorescence methods after applying one of many available labeling techniques. Polymeric nanoparticles can be labeled by covalently functionalizing the polymers with dyes such as Texas red32 or fluorescein isothiocyanate (FITC)34,39 or by loading the polymeric NP with a fluorescent dye such as 6-coumarin.30,40 Covalently attached FITC has also been used to label fullerenes,41 multi-walled nanotubes (MWNTs)42 and single-walled nanotubes (SWNTs)43 to monitor uptake. Fluorescently labeled iron oxide nanoparticles have been prepared by conjugation of biotin and labeling with streptavidin-FITC35 or by formulating with dipyrromethene boron difluoride linked polylactic acid.31 Gold NPs have been labeled by Texas red-modified protein adsorption23 while mesoporous silica nanoparticle uptake has been monitored by co-condensation with FITC44 or rhodamine B45 for labeling. While these labeling schemes facilitate fluorescence quantitation, the presence of surface dyes or incorporated species may alter the chemical or physical properties of the nanoparticles, thereby altering the biological implications of their exposure. Therefore, these studies require additional control experiments to account for affects of the fluorescent surface dyes or incorporated species.
2.4 Novel uptake analysis techniques
A variety of novel techniques have recently been used to investigate nanoparticle uptake by cells or tissue. Feldheim and co-workers used video-enhanced differential interference contrast (VEDIC) microscopy to determine trajectories and localization of gold NPs as small as 20 nm in diameter within cells without adding a fluorescent label (Fig. 2).46 The high scattering efficiency of gold nanotubes in the near IR has also been exploited to characterize their uptake in HeLa cells.47 Additionally, two-photon luminescence microscopy has been used to monitor gold nanorod uptake and trajectories with excellent time resolution.48 The loading of superparamagnetic iron oxide nanoparticles has been found to correlate with cellular velocity distributions when placed in a magnetic field, and this correlation has been used to determine the uptake of these nanoparticles.49 The unique challenges of characterizing nanoparticle uptake into biological systems continue to drive interesting analytical research.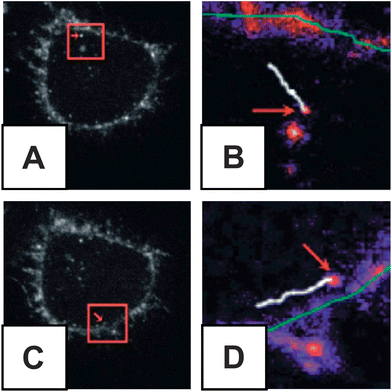 | ||
| Fig. 2 VEDIC obtained trajectories of peptide conjugate QDs within HeLa cells. (A) Cell shown with higher magnification inlay (B) showing trajectory vesicle containing QDs over 2 seconds of acquisition (white line) relative to plasma membrane (green line). (C) A second cell is shown with its inlay (D) showing QD trajectory over 1.74 seconds. (Reprinted with permission from ref. 38. Copyright 2007, American Chemical Society.) | ||
3. In vitro toxicity assessment
In vitro toxicological assessment is a vitally important tool for nanotoxicology. Compared to in vivo studies, in vitro studies benefit from being faster, lower cost, allowing greater control, and minimizing ethical concerns by reducing the number of laboratory animals required for testing. However, the extension of results from in vitro experiments for the prediction of in vivo toxicity is problematic due to the in vitro exposure conditions which usually feature much higher concentrations and exposure times than found in the cellular environment in vivo. The most commonly used in vitro assessment techniques, generally, either evaluate viability (live/dead ratio) or toxicity mechanism as summarized in Table 1. Herein, the major viability based assays are organized into the categories of proliferation, necrosis or apoptosis, and the major toxicity mechanism analyses are categorized as either oxidative stress or DNA damage detection techniques. The categorical separation of these assays is somewhat artificial as there is significant overlap between the assays. For example, DNA damage can be associated with apoptosis and oxidative stress in addition to arising directly from nanoparticle contact in the nucleus. This review is not intended to be an exhaustive review of toxicological methodology, rather it is a review of those techniques currently applied to nanotoxicology and alternative techniques available.| Assay type | Category | Cellular property/process probed | Assays | Examples of use |
|---|---|---|---|---|
| Viability | ||||
| Proliferation | Metabolic activity | MTT, XTT, WST-1, Alamar Blue | 7,51–58,69–72,74,81 | |
| DNA synthesis | [3H]Thymidine incorporation | 75,76 | ||
| Colony formation | Cologenic | 79 | ||
| Necrosis | Membrane integrity | LDH, Trypan Blue, Neutral Red, propidium iodide | 39,43,80–82,84,87–89 | |
| Apoptosis | Membrane structure | Annexin-V | 33,84,87,93 | |
| LIVE-DEAD | Esterase activity/membrane integrity | Calcein acetoxymethyl/ethidium homodimer | 51,90,91 | |
| Mechanistic | ||||
| DNA damage | DNA fragmentation | Comet, CSE | 87,95 | |
| DNA double-strand breakage | TUNEL | 96,97 | ||
| Oxidative stress | Presence of reactive oxygen species (ROS) | DCFDA, Rhodamine123 | 54,105,120,126,127,129,142 | |
| Lipid peroxidation | C11-BODIPY | 114 | ||
| Lipid peroxidation | TBA assay for malondialdehyde | 114,119,121,136–138 | ||
| Presence of lipid hydroperoxides | Amplex Red | 140 | ||
| Antioxidant depletion | DTNB | 114,142 | ||
| Superoxide dismutase (SOD) activity | Nitro blue tetrazolium | 142 | ||
| SOD expression | Immunoblotting | 142 | ||
3.1 Proliferation assays
Cellular reduction of tetrazolium salts to produce formazan dyes is the most widely used in vitro nanotoxicity assessment. The production of formazan-based dyes is monitored by optical absorbance as a measurement of cellular metabolism, which is used to assess the percentage of metabolically active cells. The most popular of these tetrazolium salts is 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT),50 which has been used to assess the in vitro toxicity of a wide variety of nanostructures.51–59 This technique has many advantages when compared to other toxicity assays because it requires minimal physical manipulation of the model cells and yields quick, reproducible results requiring simple optical density acquisition.60 However, the interpretation of these assays can be misleading as the previously held assumption that the metabolic reduction of tetrazolium salts is coupled to in vitro proliferation has been found untenable.60 Furthermore, the cellular mechanism for reduction of tetrazolium salts remains unclear as the location of the reduction has been found to be outside the mitochondria.61–63 Additionally, changes in media pH64 and culture media additives such as serum,65,66 cholesterol,67,68 or ascorbate69 have been found to alter these measurements. In assays that produce insoluble formazan dyes (such as the MTT assay), exocytosis of the crystalline product can skew results, therefore assays that produce soluble dyes (such as XTT or WST-1) are preferred. Of particular importance to nanotoxicology, interactions between nanomaterials and either the reactants or products of tetrazolium-based assays have led to spurious results as evident in experiments with SWNTs,70–72 porous silica73,74 and carbon black NPs.75 Alternative metabolic assays include [3H]thymidine incorporation and the Alamar Blue (also called resazurin) reduction assay.72,76 Uptake of [3H]thymidine into newly synthesized DNA is a highly sensitive measurement of cellular proliferation with a wide dynamic range but is often avoided due to its relatively high price and in vitro toxicity.77,78 Alamar Blue has been relatively recently applied to nanotoxicological studies by assaying cellular redox potential.76 Alamar Blue is reduced to a soluble fluorescent product, resorufin (λem = 590 nm), providing simpler sample preparation compared to the MTT assay. However, interpretation of Alamar Blue results is difficult because the biochemical mechanisms of Alamar Blue reduction have not yet been explored. Additionally, nanoporous silicon has been found to react with Alamar Blue in the absence of cells.74 Interactions between nanomaterials and probe molecules can be avoided altogether through the use of cologenic assays,79,80 where colonies of highly proliferating cells are counted by visual inspection after exposure to nanomaterials.723.2 Necrosis assays
Membrane integrity is another cellular characteristic commonly used to determine viability during in vitro nanotoxicology experiments.39,43,51,81–85 Typical methods for assessing membrane integrity either monitor (1) the uptake of a supravital dye, such as Trypan Blue (TB),39,81,86 Neutral Red (NR),39,82,83,87 or propidium iodide (PI)43,84,88 or (2) the leakage of active enzymes into the cell media using the LDH assay.85,89,90 TB and PI are charged molecules excluded from live cells. Cells with ruptured membranes allow for the passage of TB into the cell where it stains with a strong absorption at ∼605 nm. When PI enters a cell, it intercalates into DNA and dsRNA where it fluoresces at 617 nm. NR is uncharged at physiological pH and is able to enter both live and dead cells. In live cells it is protonated in the highly acidic lysosomes (pH ≈ 4.8) where NR accumulates and absorbs strongly at 540 nm. These techniques provide highly reproducible results and can have increased throughput by combining with a flow cytometry technique. Additionally, a combination calcein acetoxymethyl ester/ethidium homodimer assay can be used to label both living and necrotic cells in nanotoxicology52,91,92 where calcein acetoxymethyl ester undergoes enzymatic hydrolysis by active esterases within living cells, producing a green fluorescent product and ethidium homodimer, a red fluorescent dye, accumulates only within dead cells.933.3 Apoptosis assays
The aforementioned membrane dye exclusion techniques are only capable of detecting necrosis or late stage apoptosis conditions of cells and are often combined with apoptosis detection assays to form a more complete picture of cell death. Apoptosis assays have been used extensively during nanotoxicological research and include inspection of morphological changes, the annexin-V assay,15,85,88,94 DNA laddering,95 the Comet assay88,96 or the TUNEL assay.97,98 Inspection of morphological changes during apoptosis is the least instrumentally intensive method for characterizing apoptosis, requiring only a light microscope and visual inspection. Despite the low cost, this method is not used widely in nanotoxicology due to the time-intensive nature of the method, and possible misidentification arising from confusion of apoptotic cells with endocytotic engulfment of apoptotic bodies. The annexin-V assay, as seen in Fig. 3, takes advantage of apoptotic restructuring of the plasma membrane that exposes phosphatidylserine on the exterior of the cell.99 Annexin-V is a phosphatidylserine-specific binding substrate that is excluded from the interior of living cells. When labeled with a fluorescent tag, such as FITC, this binding can be used to fluorescently label the membranes of early and late stage apoptosis in cells. Enzyme-mediated intranuclear DNA fragmentation is an identifying characteristic of apoptosis100,101 so DNA fragmentation is often assayed as an assessment of apoptosis.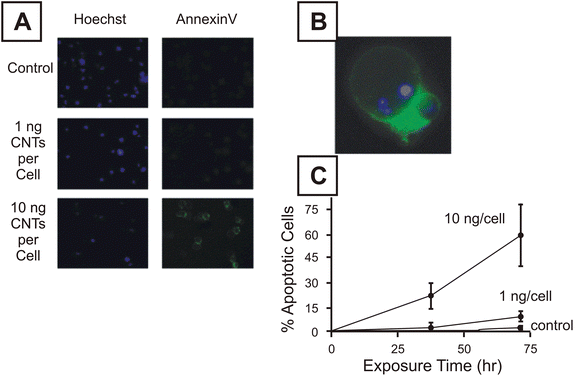 | ||
| Fig. 3 Results of annexin-V assessment of apoptosis as a result of T cell exposure to carbon nanotubes. (A) Hoechest (DNA labeling) dye (left) stained and annexin-V stained cells after exposure to 0, 1 and 10 ng/cell oxidized carbon nanotubes. (B) Higher magnification of apoptotic cells stained with annexin-V. (C) Graphical representation of time course exposure of cells to carbon nanotubes between 0 and 10 ng/cell. (Reprinted with permission from ref. 94. Copyright 2006, Elsevier.) | ||
3.4 DNA damage assays
DNA damage in the form of unrepaired single- and double-strand DNA breaks, can occur from nanoparticle exposure via changes in the oxidative environment, apoptosis, or by physical interactions between the DNA and nanoparticle, resulting in what is termed genotoxicity. DNA laddering, the oldest DNA damage assay technique, characterizes this fragmentation by isolating and fluorescently labeling DNA from cells exposed to a potential toxicant in culture. DNA damage is then detected by gel electrophoresis. DNA damage is most commonly determined by use of the Comet assay, also called the single-cell gel electrophoresis (SCGE) assay. In the Comet assay, originally described by Singh et al., one embeds the cells in agarose gel, lyses the cells and denatures the DNA, which is then subjected to labeling with ethidium bromide (for quantitation) and electrophoresis (for separation).102 The amount of DNA damage is indicated by the amount of tailing, or DNA fragments trailing after electrophoresis. Numerous studies have used the Comet assay to determine DNA damage upon nanoparticle exposure.103–109 Recent work by Barnes et al. has shown that the Comet assay showed no genotoxicity at the TiO2 nanoparticle concentrations tested (4 and 40 µg/mL).104 While not focusing on the toxicity of the TiO2 nanoparticles, they were able to show that the use of a standard protocol for in vitro testing can be performed both quantitatively and reproducibly using immortal 3T3 cells and supplied TiO2 nanoparticle (both purchased and synthesized in-house). While this study indicates that the Comet assay can be reproducible, the authors also note that strict adherence to the sample preparation and handling was necessary to achieve concordance. Therefore, while it is possible to achieve reproducibility, it is likely that handling differences will complicate the interpretation and comparison of the genotoxic effects of nanoparticles. Another DNA damage assay, the TUNEL assay, which derives its name from ‘TDT-mediated dUTP-biotin nick-end labeling’,110 relies on double-strand breakage, like the damage necessary for DNA fragmentation during apoptosis. The most popular method for this assay uses exogenous DNA polymerase I to repair isolated fragmented nuclear DNA. The polymerase is added with 5-bromo-2-deoxyuridine, a synthetic deoxythymidine analogue, to incorporate BrdU into repaired double-strand breaks in apoptotic cells.111 A FITC conjugated anti-BrdU antibody is used to label BrdU-containing DNA to detect the amount of double-strand breaks in nuclear DNA.3.5 Oxidative stress
In a cellular environment, engineered nanomaterials may disturb the oxidative balance of the cell. This phenomenon is called oxidative stress and it results in abnormally large concentrations of intracellular reactive oxygen species (ROS) such as superoxide (O2−), hydroxyl radical (HO˙), peroxy radical (ROO˙), and hydrogen peroxide (H2O2); or reactive nitrogen species (RNS) such as nitric oxide (NO˙), peroxynitrite anion (ONOO−), peroxynitrous acid (ONOOH) and nitrosoperoxycarbonate anion (ONOOCO2−). The generation of abnormally large concentrations of ROS and RNS can have many toxicological implications by reaction with proteins, lipids or nucleic acids, leading to abnormal cellular function.112–114 Studies in oxidative stress resulting from nanoparticle exposure have been extensive.9,13,71,85,106,115–121 The probe molecule 2,2,6,6-tetramethylpiperidine (TEMP) reacts with O2− to form a stable radical (TEMPO) that can be detected directly using X-band electron paramagnetic resonance (EPR).122 However, this technique is typically avoided due to the relatively high cost of EPR instrumentation. Alternatively, fluorescent probe molecules reactive to ROS/RNS are widely used to assess ROS/RNS production in vitro;123 however, mechanistic interpretation of the results from these assays can be misleading because the probes are reactive with a wide variety of reactive species.124 Amongst the most widely used of these probes in nanotoxicology is 2′,7′-dichlorofluorescein diacetate (DCFDA),125 a non-fluorescent probe that is able to cross the cell membrane. Within the cell DCFDA is hydrolyzed by esterases, converting it to 2′,7′-dichlorofluorescein, a probe that is reactive to HO˙, ROO˙, RO˙ and H2O2 (in the presence of cellular peroxidases). Therefore, this is a general test for determining oxidative stress through the production of 2′,7′-dichlorofluorescein as a fluorescent product (λex 512 nm/λem 530 nm).55,71,106,126–129 Oxidative stress measurements using DCFDA should be used cautiously when apoptosis occurs as a result of nanoparticle exposure. Cytochrome c, released from the mitochondria during apoptosis, has been found to be a strong oxidative catalyst for DCFDA130 and could lead to exaggerated measurements of oxidative stress. Dihydrorhodamine123 is also used as a general oxidative stress probe131 as it is oxidized by ROS and RNS to Rhodamine123 which fluoresces (at λex 507 nm/λem 529 nm) and has been applied to nanomaterials.71,121 Dihydrorhodamine123 is insensitive to O2˙−- or NO˙-mediated oxidation.132 Oxidative stress probes designed to localize within specific regions of the cell are used to provide insight on the source and action of ROS and RNS during NP-mediated toxicity. Hydroethidine is a non-fluorescent probe selective for superoxide via an oxidative fluorescent product. This is commonly used under the commercial name MitoSOX Red, where hydroethidine is modified with hexyl triphenylphosphonium, to target and detect superoxide production within the mitochondria (λex 510 nm/λem 580 nm)133,134 and has been used to characterize oxidative stress caused in neurons by titanium dioxide nanoparticles.117,129 To visualize lipid peroxidation, C11-BODIPY was developed as a ratio-fluorometric dye that localizes within the plasma membrane.135 Non-selectively, the dye fluorescence shifts from (λex 581 nm/λem 610 nm) to (λex 485 nm/λem 510 nm) when oxidized and has been used to characterize lipid peroxidation by nano-fullerenes as seen in Fig. 4.115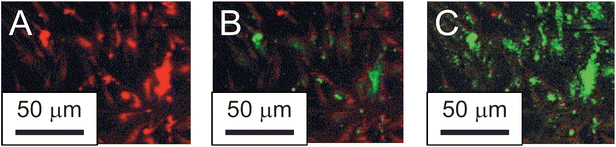 | ||
| Fig. 4 Lipid peroxidation detected by C11-BODIPY as a result of (A) 0 min, (B) 30 min, and (C) 90 min exposure to nano-C60. (Reprinted with permission from ref. 115. Copyright 2005, Elsevier.) | ||
Additionally, oxidative stress can be determined by biomarker stress products from lipid and protein peroxidation, or DNA fragmentation. Lipid peroxidation has been used to monitor nanotoxicity by the thiobarbituric acid assay of malondialdehyde, the end product due to peroxidation of polyunsaturated fatty acids, when cells were exposed to nanomaterials.115,120,136–138 It is important to note that there is as of yet no universal biomarker for oxidative stress.124 Oxidative stress can also be examined via the detection of lipid hydroperoxides139 by fluorescent labeling with N-acetyl-3,7-dihydroxyphenoxazine (Amplex Red) followed by HPLC separation with fluorescence detection. Although not extensively explored as a biomarker of oxidative stress, the depletion of cellular stores of dopamine by ROS and the increase in oxidative products after exposure to nanomaterials has been characterized by HPLC-EC.140
Other oxidative stress biomarkers include upregulation of cellular compensation mechanisms as evident by alterations in superoxide dismutase or glutathione production. Increases or decreases in these responses can be interpreted as evidence for oxidative stress as the cell either compensates for increased stress by upregulating the production of antioxidants or the exhaustion of cellular stores of superoxide disumutase (SOD) or glutathione (GSH) by oxidation from RNS or ROS. GSH is an essential antioxidant that is oxidized during oxidative stress to form a GSH-GSH disulfide between two GSH molecules yielding GSSG. The most quantitative assessment monitors the ratio of GSH and its disulfide oxidative product GSSG using HPLC;141 however, the derivatization and chromatographic separation steps are time-consuming and allow for auto-oxidation, leading to over-estimation in the amount of GSSG. For this reason, combined GSH and GSSG have been assayed instead during the nanotoxicology studies to date, using 5,5′-dithio-bis(2-nitrobenzoic acid) (DTNB).115,142 DTNB is used in the presence of NADPH, where NADPH fuels the reduction of GSSG to GSH by glutathione reductase; the resulting total GSH concentration is determined by the colorimetric detection of 5-thio-2-nitrobenzoic acid (λabs 412 nm) after reaction of DTNB with GSH.143 There are alternative assays for determining individual concentrations of GSH144 or GSSG145 but these are avoided due to their lack of sensitivity as they are approximately 3 orders of magnitude less sensitive. SOD is an antioxidant enzyme in which two major isoforms, SOD-1 and SOD-2, catalyze superoxide to form hydrogen peroxide and molecular oxygen. SOD has been characterized both in terms of expression142 and activity. SOD activity is determined indirectly via the inhibition of superoxide oxidation of a colored substrate, nitro blue tetrazolium, where superoxide is generated via exogenous xanthine-xanthine oxidase.146 SOD-1 and SOD-2 expression is characterized by immunoblotting techniques where, after isolation and electrophoretic separation, they are tagged with commercially available primary antibodies, then with secondary antibodies conjugated to reporter enzymes – in this case, horseradish peroxidase, whose activity can be characterized colorimetrically, fluorimetrically or chemiluminescently.
3.6 Novel in vitro toxicological techniques
Recently, a number of new approaches have been applied to characterizing in vitro nanotoxicity. Gene expression analysis has been recently applied to the study of nanotoxicity.22,147 This technique compares labeled RNA collected from nanoparticle-exposed and control cells through a competitive binding assay, using commercially available microarrays of human cDNA libraries. This yields a large table showing both increased and decreased RNA expression as a result of nanoparticle exposure, although the toxicological implications of such changes in expression are often unclear. The high-content screening assay (HSA) is a tool that has proven to be useful for screening toxicity in potential drug applications148,149 and has been recently applied to nanomaterials.147,150 HSA is a high-throughput technique that uses automated, commercially available instruments to perform computer-assisted microscopic image analysis. HSA examines the morphology of exposed cells along with fluorescent characterization, using the aforementioned fluorescent probes, to assess multiple toxicity modes simultaneously (i.e. apoptosis, necrosis, oxidative stress, etc.). In another recent paper, microelectrodes were used to probe the effects that nanoparticles had on the release of electroactive molecules from single-cells.151 This technique allows for individual cell analysis in mixtures of cell types and elucidates changes in messenger molecule concentrations as well as alterations in the biophysical properties of exocytosis. However, the full toxicological ramifications of such messenger concentration and biophysical changes are unclear.4. In vivo toxicity assessment
Another course of study for nanotoxicity, beyond the in vitro assays, is to study the impact of nanoparticle exposure within intact living organisms. These in vivo studies are typically undertaken in mice152–155 or rats.156,157 While in vivo studies bear ethical consideration, due to the use and sacrifice of animals, these studies allow examination of long-term effects, tissue localization, biodistribution, and retention/excretion. These types of in vivo studies can inform the choice of relevant model system for further in vitro studies, as well as provide toxicity information not available through in vitro study. Beyond the simple LD50 (lethal dose for 50% of participating animals) study, in vivo nanoparticle toxicity studies typically focus on one or more of three major areas: changes in blood serum chemistry and cell population, changes in tissue morphology, examined using histology, or the overall nanoparticle biodistribution. Typically, in vivo experiments utilize several of these, and other, techniques to maximize the toxicity information gained. As with in vitro studies, one must consider many aspects prior to beginning studies, such as nanoparticle choice and characterization, prior to exposure, and choice of relevant model system. Additionally, one must also consider issues of exposure route, nanoparticle dosing and specific tissue/organ or proteins/cell type of interest, to examine histological or serum/hematological changes, respectively. While there are many potential model systems, serum markers, and tissues to examine, the examples described herein, though not exhaustive, represent specific choices that are commonly studied in nanotoxicology.4.1 Biodistribution and clearance
Biodistribution and clearance studies usually occur in tandem to examine the overall process of tissue or organ localization and follow the passage of the nanoparticles through the animal over time. There are several reviews which describe the impact that nanoparticle properties, such as particle composition, size, core-shell composition and surface functionality/charge, can have on tissue distribution and clearance rates.158,159 Nanoparticles are detected within live or recently sacrificed whole animals and fixed tissues using fluorescence (native or tagged) or scintillation (via radiolabels), and ICP-MS or ICP-AES (using homogenized tissue samples).156,160–164 Additionally, there can be gross examination of morphological changes for whole organs as shown in Fig. 5.165 Another subset of biodistribution studies examines the efficacy of tumor-targeted nanoparticles in vivo,154,156,166 as shown in Fig. 6.167 Clearance studies are accomplished by long-term study of the metabolism and excretion of the nanoparticles at various time points after exposure. This is largely done by examining blood samples or sacrificed animals throughout the study to determine the amount of nanoparticle present.168–170 The method of nanoparticle quantitation, using ICP-MS, radiolabeling and fluorescence, is dependent on the type/properties of nanoparticle being studied. Nanoparticle biodistribution and clearance studies provide insight into nanoparticle localization, retention and passage from animals. However, since many of the widely used quantification techniques require the addition of a radiological or fluorescent tag, the presence of these tags may alter the biodistribution and clearance of these nanomaterials. There remains a great need for rapid analytical methods for the detection and localization of nanomaterials in vivo.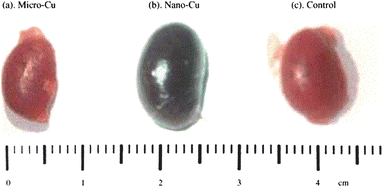 | ||
| Fig. 5 During in vivo biodistribution studies, organ morphology is often examined. In this example, excised kidneys from mice after copper exposure with (a) 17 µm diameter particles, (b) 23.5 nm diameter particles, and (c) control conditions are shown. (Reprinted with permission from ref. 165. Copyright 2006, Elsevier.) | ||
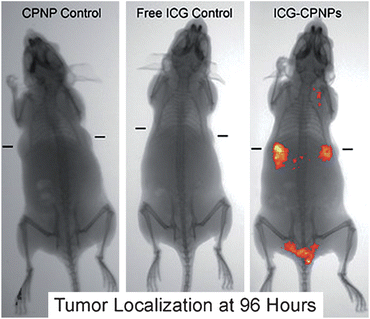 | ||
| Fig. 6 Biodistribution of indocyanine green labeled tumor-targeted nanoparticles, post-exposure for 96 hours using unlabeled calcium phosphate nanoparticles (CPNP control), free indocyanine green (Free ICG Control), or indocyanine green labeled nanoparticles (ICG-CPNPs). The implanted targeted tumor location is denoted by dash marks in each image. (Reprinted with permission from ref. 167. Copyright 2008, American Chemical Society.) | ||
4.2 Hematology and serum chemistry
One of the most common in vivo toxicity assays is to examine blood composition, both cell type and serum chemistry, for changes which occur after exposure to nanoparticles. Here the homeostasis of the blood is used as a proxy for toxicity, where any deviation, either increase or decrease in blood constituents, from the normal (pre-exposure) condition is representative of toxicity. In the case of hematology, cell population, from either general consideration of cells such as red and white blood cells or specific consideration of cells such as T cells and macrophages, and distribution are considered during these studies. Both hematology and serum chemistry studies are typically performed on automated analyzers, such as those available from Abbott, Beckman or Roche, after collection of whole blood. Cell populations are typically distinguished using volume cutoffs to indicate cell type, such as platelet or red blood cell, which are sorted after volume classification and counted, measured as changes in resistivity through an aperture, similar to the counting and sizing done in flow cytometry. The blood sample can be from live animals after exposure, or from recently sacrificed animals. In one study, Baker et al. found that for rats exposed to inhaled C60 there was a decrease in the overall red blood cell population when using 55 nm diameter C60 aggregates and a decrease in white blood cell count when the aggregate size was changed to 930 nm.157 In combination with multiple in vivo assessments, including histology of various organs, behavioral observation and bronchoalveolar lavage, they concluded that there was minimal toxicity for the 55 nm diameter C60 nanoparticle aggregates. During serum chemistry studies, perturbations in serum chemistry levels are examined, but generally are reported in a qualitative manner (as increased or decreased levels of the serum analyte of interest). During automated studies of serum chemistry, protein levels are generally quantitated by use of either turbidity or nephelometric measurements, where antibodies for the proteins of interest are added to the sample, causing aggregation and altered light transmission/scattering. Alternatively, serum proteins can also be separated, using capillary electrophoresis, and quantitated, using a variety of protein-specific labeling methods (i.e. fluorescence). Some of the typical biochemical indicators examined are: alanine aminotransferase, aspartate aminotransferase, total bilirubin, alkaline phosphatase, albumin, creatinine, hemoglobin, glucose, urea nitrogen, and total protein.155,157,161,169,171 In one study, Huang et al. examined the impact Au-Au2S core-shell nanoparticles had on serum levels, measuring nine different biochemical indicators, between three and 14 days post-exposure.169 In combination with their other in vivo studies, they found that there were no statistical differences for any of the biochemical indicators examined though they found that there were other indications of toxicity after exposure. Because it is important for the cell populations and serum chemistry levels to be highly regulated, it is necessary to understand how any alterations indicate toxicity and to examine these changes in a dynamic fashion over the course of the exposure and post-exposure. Additionally, increasing the temporal resolution between sample collections may provide insight into the dynamic impact of nanoparticle exposure over shorter timescales.4.3 Histology/histopathology
Another widely utilized in vivo assay is the examination of histological changes for given cells/tissues/organs after exposure to nanoparticles. Histological examination is performed on tissues that have been fixed after sacrifice of the exposed animal, and changes in the tissue and cell morphology are assessed using light microscopy. The standard procedure for histological preparation is to fix the tissue/organ, which is then embedded into paraffin and sectioned via microtome into thin sections. These thin sections can then be stained with dyes, most commonly the H & E stain or hematoxylin (stains the nucleus/nucleic acids blue) and eosin (stains the cytoplasm pink) stain, as shown in Fig. 7.172 Staining provides image contrast, in order to highlight the changes in cell or overall tissue morphology, as measured visually. Though this technique has been used extensively since the 19th century it has limitations, most importantly, the possibility of artifacts being induced by the process of fixation, embedding, sectioning or staining. Some of the typical tissues examined after nanoparticle exposure include: brain, eyes, lung, liver, kidneys, spleen, and heart.152,155,157,173–175 In one example, Lin et al. examined the histopathology and biodistribution of the commercially available quantum dot, QD705, to develop a theoretical model to predict tissue distribution and retention in mice.170 They examined the QD localization, tissue distribution and metabolism/excretion over 6 months following IV exposure. The authors found that though there was little evidence of pathology, seen with light microscopy, there was tissue damage when examined by TEM that was in agreement with their other findings. Histological/histopathological assays provide physical evidence of morphological changes, which assumes that toxicity correlates with changes in tissue and cell morphology of a scale that can be visualized using light microscopy.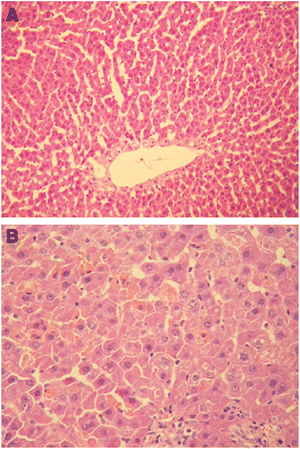 | ||
| Fig. 7 Histopathology images of rat liver tissue, using H & E staining, 6 days post-exposure where (A) control rats show normal tissue morphology and (B) scattered dot necrosis was seen in rats exposed to nano-copper at 200 mg/kg/d. (Reprinted with permission from ref. 172. Copyright 2008, Elsevier.) | ||
4.4 Novel in vivo assays
In vivo studies are complicated by the challenge of examining what is occurring upon and shortly after exposure, where quantitation in a living system prevents facile assessment of important parameters, such as the localized concentration and dynamic trafficking of nanoparticles. To overcome the challenges specific to nanoparticle toxicity studies, new techniques are being used in conjunction with the aforementioned in vivo techniques or on their own. Additionally, in vivo nanoparticle toxicity studies could benefit from dynamic in vivo assays, where intact functioning animals are used instead of fixed or highly processed samples. The use of common bioanalytical techniques where in vivo application is routine, such as with microfluidics,176–179 and microelectrochemistry,180–182 could be applied to assess nanoparticle exposure. In these microfluidic and microelectrochemistry studies, samples are taken directly from freely moving animals through an implanted probe. This allows dynamic in vivo measurements which are limited by the sampling and detection rates of the probes and detectors, respectively. Additionally, work by Tsai and co-workers has utilized an automated blood sampling system for use with freely moving animals as well.171,183 These types of dynamic sampling methods would overcome some of the limitations of the commonly used in vivo assays where measurement usually provides a static picture which may be skewed by artifacts induced by sample handling and preparation.5. Roles for analytical chemists in nanotoxicology
Analytical chemists are particularly suited to address the analytical challenges in nanotoxicology because they are accustomed to developing new technology, pushing towards lower limits of detection, and navigating complex samples. One appropriate challenge for analytical chemists in nanotoxicology is in the area of nanoparticle characterization. Most of the current techniques used for physical nanoparticle characterization give only limited information before or after engineered nanoparticles are in the biological environment. Commonly used methods like electron microscopy, scanning probe microscopy, and dynamic light scattering reveal nanoparticle size and distribution but give no information about aggregation/agglomeration that may occur within the biological environment, adsorption of biomolecules, or degradation of the nanoparticle itself, all of which may change during residence within the biological system and would greatly influence nanoparticle uptake and/or behavior. Nanotoxicologists would certainly benefit from technological advances to assess the extent of nanoparticle aggregation in vitro or in vivo or from the creation of accurate bioenvironment models to perform these measurements using existing instruments.It is possible to imagine many routes to monitor nanoparticle-adsorbed surface species. Mass spectrometry (MS) approaches could be useful for identifying unknown species adsorbed on nanoparticles but require volatilization technique development. There are a few recent examples of nanoparticle-enabled MS that, though not explicitly linked to nanoparticle toxicity studies, demonstrate the potential of this approach. For example, Kong et al. used carboxylated/oxidized diamond nanoparticles to extract proteins from blood, centrifuged the nanoparticles, mixed them with a MALDI matrix, and performed MS analysis with 2 orders of magnitude improvement in sensitivity over analysis done without nanoparticles.184 Similarly, Chang et al. employed oleate-modified iron oxide nanoparticles to capture peptides and proteins and collected them magnetically before mixing them with MALDI matrix and achieving spectra with minimized interference and suppression.185 Spectroscopic techniques, such as surface-enhanced Raman scattering from noble metal nanoparticles, have the potential to allow dynamic assessment of adsorbed species in vitro or in vivo but need more efficient photon collection methods than are commonly available. Again, there are some recent literature precedents, unrelated to nanoparticle toxicity, that prove the potential utility of this approach. Stuart et al. installed a SERS-active nanostructured substrate and imaging window inside a rat and used partition layer-assisted SERS to monitor real-time in vivo glucose approach to the nanostructured substrate.186 Meanwhile, Kneipp and co-workers used nanoparticles taken up into lysosomal compartments to monitor small molecule species and pH changes in vitro.187,188 As nanoparticle design gets more advanced and more components are incorporated into a single engineered nanostructure (e.g. multifunctional drug delivery nanoparticles),152,189 it will be increasingly necessary to dynamically characterize nanoparticle degradation to determine if unintended nanoparticle components are leaching into the biological environment. In a related challenge, one major concern among nanotoxicologists is the accidental exposure of manufacturing or research employees to nanoparticulate material.3 While current technology is capable of detecting the presence of even small numbers of nanoparticles, there is not an inexpensive, portable version of this technology that could be installed on each lab bench or fabrication line to warn workers when excessive nanoparticle exposure is occurring. This is clearly a challenge that could most efficiently be addressed by analytical aerosol scientists.
The wide variety of in vitro and in vivo assays that are employed to assess nanoparticle toxicity are used because these are the tools that were already available for molecular toxicology when nanoparticle toxicity questions arose. The first major problem with using these assays to assess nanoparticle toxicity is that the modes of nanoparticle toxicity might not be the same as those incurred by molecular toxicants. Cellular uptake and localization of nanoparticles will almost certainly be different than internalization of molecular species and these will likely lead to different modes of toxicity. While there are some examples in the literature exploring nanoparticle uptake and localization, this is currently far from well understood and, apparently, varies significantly based on the nanoparticle being used. For example, while Lühman et al. demonstrated that block copolymer nanoparticles enter cells using a variety of pathways and localize in the cytoplasm,190 Li et al. showed that fullerenes are taken up through clatherin-mediated endocytosis and are associated with lysosome-like vesicles,41 and Ryan et al. found peptide-targeted gold nanoparticles localized in the nucleus.24 These differences in uptake and localization present two important challenges in the area of technological development. First, the classical tools used to assess nanoparticle uptake and localization (TEM,191 emission spectroscopy,24 confocal fluorescence imaging,30etc.) do not allow dynamic measurement with unlabeled nanoparticles, and new technology development in this area would give important insight. Total internal reflectance fluorescence (TIRF) has great potential for real-time imaging of fluorescent nanoparticles as in the work by Lee et al. examining uptake and localization of dendrimer nanoparticles for gene delivery.192 Dark field scattering looks promising for nanoparticles with large scattering cross-sections, like noble metal nanoparticles, as in the work by Xu and co-workers tracking the real-time uptake of silver nanoparticles into zebrafish embryos.85 The second scientific challenge for analytical chemists based on cellular uptake and localization of nanoparticles is to create assays that are sensitive and selective for the biomarkers that indicate toxicity. Unfortunately, the varied cellular localizations lead to varied modes of toxicity and thus some confusion about the appropriate biomarkers; while nuclear localization may lead to direct DNA damage, acidic organelle localization is more likely to initiate the release of toxic ions during nanoparticle degradation. Clearly, different modes of toxicity and possible downstream influence of one cell type on another require assays that measure different (and perhaps multiple) species, and tackling this challenge will require close cooperation between toxicologists and analytical chemists. Once toxicologists have discovered a relevant toxicity mechanism for a nanoparticle or class of nanoparticles of interest, analytical chemists can work to develop assays that are both sensitive to and selective for this species or cell function.
The second major problem with using traditional toxicology assays is that nanoparticles may interfere directly with the signal transduction based on the increased reactivity of their surface sites compared to bulk materials. In one example, Ag nanoparticles were found to interact directly with the tetrazolium salt of the MTT assay, suggesting greater than 100% viability of nanoparticle-exposed cells based on the ability of the nanoparticles to generate the colored formazan product.73 Even with results like this, tetrazolium salt-based assays are still the most commonly used methods in nanoparticle in vitro toxicity studies. This field would benefit greatly if the basic tenets of analytical chemistry were applied, creating assays that are (1) specific for signals that indicate important toxicity mechanisms, (2) sensitive to these signals with an appropriate dynamic range, (3) measuring signals from specific and known cells or tissues, and (4) monitoring real-time toxicity response, all with appropriate positive and negative controls. The first and second of these desirable characteristics are discussed in the previous paragraph. The third requires either spatial resolution in a measurement sufficient to distinguish one cell or tissue from another or single-cell measurements instead of ensemble-averaged measurements. The fourth characteristic of real-time, dynamic response is critical for understanding the half-life of nanoparticles in a biological system and effects related to nanoparticle dwell time. In many cases, cell washout is possible, leading to many important questions such as: do toxic effects disappear when the nanoparticles depart a cell or tissue; and, how long does it take for a cell to return to normal behavior after nanoparticle expulsion? Finally, appropriate control experiments must be performed with each assay. There have been multiple examples in the literature where nanoparticles were originally thought to be toxic to a particular cell or tissue but later studies demonstrated that the actual toxicant was a solvent, surfactant, or ion in solution.12,42
The goal of using in vitro nanoparticle toxicity assays is to predict in vivo toxicity. Unfortunately, there is often little correlation between the two, with in vitro toxicity assays predicting more severe cell damage than a whole animal experiences. To better correlate in vitro and in vivo measurements, it is critical to develop accurate dosing concentration representations, procedures, and assessments;193 clearly, this is another area where the natural strengths of analytical chemists could be leveraged. This must be done in parallel with research in the area of nanoparticle uptake efficiency and mechanisms because correlation of in vivo and in vitro toxicity assessments is only possible if the same nanoparticle dose can be delivered in both cases. Equivalent dosing is difficult to achieve because the nanoparticle experiences a vastly more complicated delivery route and environment when administered in vivo. While the four likely routes of nanoparticle exposure are known to be ingestion, inhalation, injection, and skin penetration, the final destination of nanoparticles entering via each of these routes is unknown (Fig. 8) and likely dependent on the nanoparticles' chemical and physical characteristics.194 By far, the most complete understanding of the physiological destination of nanoparticles exists for inhaled nanoparticles;195–198 but even this case suffers from a paucity of data regarding nanoparticles composed of varied materials.
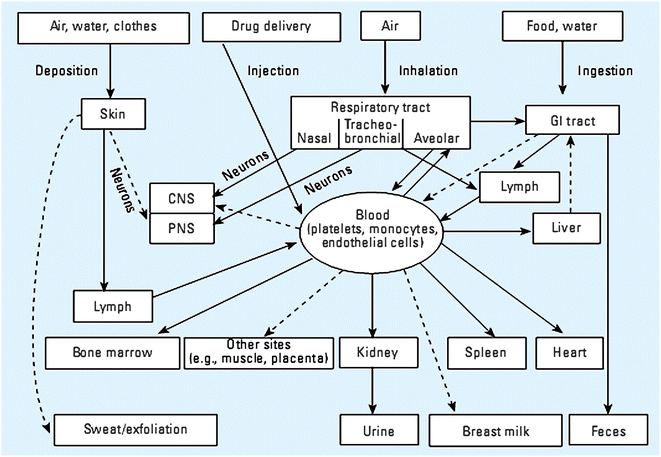 | ||
| Fig. 8 Expected and known nanoparticle exposure and clearance routes. (Reprinted with permission from ref. 194. Copyright 2005, National Institute of Environmental Health Sciences.) | ||
6. Conclusions
As nanomaterial use in both commercial goods and novel applications expands, the need for relevant, accurate and predictive nanotoxicological assessment also grows. This review details the wide variety of techniques currently being used to address nanotoxicity both for researchers working currently in the field and for scientists interested in developing new assessment techniques. Drawing conclusions from the current body of nanotoxicological work is challenging because each research group studying nanoparticle toxicity chooses a different model system, different exposure conditions, and a different measurable mode of toxicity. Additionally, there is little investigation of assay reproducibility both in terms of intra- or inter-laboratory comparison. Accordingly, it is impossible to consolidate the acquired data and look for overall trends in nanoparticle toxicity. In order to meet the overarching goal of determining a set of design rules for safe nanomaterial use based on predictive nanoparticle toxicity, standard protocols will need to be developed. These protocols will require accurate characterization of nanomaterials, high-throughput in vitro screening to identify potentially problematic nanomaterials, followed by in vivo toxicity studies based on those materials, and mechanistic studies using relevant models revealed by in vivo toxicity assessment. The output from this cycle can then be used to further refine future nanotoxicity studies and inform safe nanomaterial design. The skill set of analytical chemists uniquely qualifies them to collaboratively address challenges in designing and implementing these critically needed protocols.Acknowledgements
This work was funded by the National Science Foundation (CHE-0645041).References
- N. Lewinski, V. Colvin and R. Drezek, Small, 2008, 4, 26–49 CrossRef CAS.
- R. Brayner, Nano Today, 2008, 3, 48–55 CrossRef.
- A. D. Maynard, R. J. Aitken, T. Butz, V. Colvin, K. Donaldson, G. Oberdoerster, M. A. Philbert, J. Ryan, A. Seaton, V. Stone, S. S. Tinkle, L. Tran, N. J. Walker and D. B. Warheit, Nature (London, UK), 2006, 444, 267–269 Search PubMed.
- Q. L. Fan, K. G. Neoh, E. T. Kang, B. Shuter and S. C. Wang, Biomaterials, 2007, 28, 5426–5436 CrossRef CAS.
- J. Yang, E. J. Cho, S. Seo, J. W. Lee, H. G. Yoon, J. S. Suh, Y. M. Huh and S. Haam, J. Biomed. Mater. Res., Part A, 2008, 84, 273–280 CrossRef.
- A. Petri-Fink, B. Steitz, A. Finka, J. Salaklang and H. Hofmann, Eur. J. Pharm. Biopharm., 2008, 68, 129–137 CrossRef CAS.
- D. B. Warheit, T. R. Webb, V. L. Colvin, K. L. Reed and C. R. Sayes, Toxicol. Sci., 2007, 95, 270–280 CAS.
- J. J. Li, L. Zou, D. Hartono, C. N. Ong, B. H. Bay and L. Y. L. Yung, Adv. Mater., 2008, 20, 138–142 CrossRef CAS.
- P. Wick, P. Manser, L. K. Limbach, U. Dettlaff-Weglikowska, F. Krumeich, S. Roth, W. J. Stark and A. Bruinink, Toxicol. Lett., 2007, 168, 121–131 CrossRef CAS.
- P. V. Asharani, Y. L. Wu, Z. Y. Gong and S. Valiyaveettil, Nanotechnology, 2008, 19, 255102 CrossRef.
- A. E. Porter, M. Gass, K. Muller, J. N. Skepper, P. A. Midgley and M. Welland, Nat. Nanotechnol., 2007, 2, 713–717 Search PubMed.
- E. E. Connor, J. Mwamuka, A. Gole, C. J. Murphy and M. D. Wyatt, Small, 2005, 1, 325–327 CrossRef CAS.
- R. Shukla, V. Bansal, M. Chaudhary, A. Basu, R. R. Bhonde and M. Sastry, Langmuir, 2005, 21, 10644–10654 CrossRef CAS.
- W. J. Waldman, R. Kristovich, D. A. Knight and P. K. Dutta, Chem. Res. Toxicol., 2007, 20, 1149–1154 CrossRef CAS.
- Y. Pan, S. Neuss, A. Leifert, M. Fischler, F. Wen, U. Simon, G. Schmid, W. Brandau and W. Jahnen-Dechent, Small, 2007, 3, 1941–1949 CrossRef CAS.
- T. R. Pisanic, J. D. Blackwell, V. I. Shubayev, R. R. Finones and S. Jin, Biomaterials, 2007, 28, 2572–2581 CrossRef CAS.
- A. K. Gupta and M. Gupta, Biomaterials, 2005, 26, 1565–1573 CrossRef CAS.
- B. Ballou, B. C. Lagerholm, L. A. Ernst, M. P. Bruchez and A. S. Waggoner, Bioconjugate Chem., 2004, 15, 79–86 CrossRef CAS.
- J. Szpunar, Analyst, 2000, 125, 963–988 RSC.
- K. S. Subramanian, Spectrochim. Acta, Part B, 1996, 51, 291–319 CrossRef.
- R. N. Savage and G. M. Hieftje, Anal. Chem., 1979, 51, 408–413 CrossRef CAS.
- T. S. Hauck, A. A. Ghazani and W. C. W. Chan, Small, 2008, 4, 153–159 CrossRef CAS.
- B. D. Chithrani and W. C. W. Chan, Nano Lett., 2007, 7, 1542–1550 CrossRef CAS.
- J. A. Ryan, K. W. Overton, M. E. Speight, C. M. Oldenburg, L. Loo, W. Robarge, S. Franzen and D. L. Feldheim, Anal. Chem., 2007, 79, 9150–9159 CrossRef CAS.
- S. Patil, A. Sandberg, E. Heckert, W. Self and S. Seal, Biomaterials, 2007, 28, 4600–4607 CrossRef CAS.
- S. C. Wuang, K. G. Neoh, E. T. Kang, D. W. Pack and D. E. Leckband, Biomaterials, 2008, 29, 2270–2279 CrossRef CAS.
- E. Pawelczyk, A. S. Arbab, A. Chaudhry, A. Balakumaran, P. G. Robey and J. A. Frank, Stem Cells, 2008, 26, 1366–1375 Search PubMed.
- B. D. Chithrani, A. A. Ghazani and W. C. W. Chan, Nano Lett., 2006, 6, 662–668 CrossRef CAS.
- Z. R. Cui, C. H. Hsu and R. J. Mumper, Drug Dev. Ind. Pharm., 2003, 29, 689–700 CrossRef CAS.
- Y. Hu, J. W. Xie, Y. W. Tong and C. H. Wang, J. Controlled Release, 2007, 118, 7–17 CrossRef CAS.
- V. Bagalkot, L. Zhang, E. Levy-Nissenbaum, S. Jon, P. W. Kantoff, R. Langer and O. C. Farokhzad, Nano Lett., 2007, 7, 3065–3070 CrossRef CAS.
- E. Chnari, J. S. Nikitczuk, K. E. Uhrich and P. V. Moghe, Biomacromolecules, 2006, 7, 597–603 CrossRef CAS.
- C. W. Lu, Y. Hung, J. K. Hsiao, M. Yao, T. H. Chung, Y. S. Lin, S. H. Wu, S. C. Hsu, H. M. Liu, C. Y. Mou, C. S. Yang, D. M. Huang and Y. C. Chen, Nano Lett., 2007, 7, 149–154 CrossRef CAS.
- S. M. Hartig, R. R. Greene, G. Carlesso, J. N. Higginbotham, W. N. Khan, A. Prokop and J. M. Davidson, Biomaterials, 2007, 28, 3843–3855 CrossRef CAS.
- C. Becker, M. Hodenius, G. Blendinger, A. Sechi, T. Hieronymus, D. Muller-Schulte, T. Schmitz-Rode and M. Zenke, J. Magn. Magn. Mater., 2007, 311, 234–237 CrossRef CAS.
- A. Vogt, B. Combadiere, S. Hadam, K. M. Stieler, J. Lademann, H. Schaefer, B. Autran, W. Sterry and U. Blume-Peytavi, J. Invest. Dermatol., 2006, 126, 1316–1322 CrossRef CAS.
- O. Seleverstov, O. Zabirnyk, M. Zscharnack, L. Bulavina, M. Nowicki, J. M. Heinrich, M. Yezhelyev, F. Emmrich, R. O'Regan and A. Bader, Nano Lett., 2006, 6, 2826–2832 CrossRef CAS.
- G. Ruan, A. Agrawal, A. I. Marcus and S. Nie, J. Am. Chem. Soc., 2007, 129, 14759–14766 CrossRef CAS.
- M. Huang, E. Khor and L. Y. Lim, Pharmacol. Res., 2004, 21, 344–353 CrossRef CAS.
- J. Panyam, S. K. Sahoo, S. Prabha, T. Bargar and V. Labhasetwar, Int. J. Pharm., 2003, 262, 1–11 CrossRef CAS.
- W. Li, C. Chen, C. Ye, T. Wei, Y. Zhao, F. Lao, Z. Chen, H. Meng, Y. Gao, H. Yuan, G. Xing, F. Zhao, Z. Chai, X. Zhang, F. Yang, D. Han, X. Tang and Y. Zhang, Nanotechnology, 2008, 19, 145102 CrossRef.
- Y. Zhu, T. C. Ran, Y. G. Li, J. X. Guo and W. X. Li, Nanotechnology, 2006, 17, 4668–4674 CrossRef CAS.
- K. Kostarelos, L. Lacerda, G. Pastorin, W. Wu, S. Wieckowski, J. Luangsivilay, S. Godefroy, D. Pantarotto, J. P. Briand, S. Muller, M. Prato and A. Bianco, Nat. Nanotechnol., 2007, 2, 108–113 Search PubMed.
- Y. S. Lin, C. P. Tsai, H. Y. Huang, C. T. Kuo, Y. Hung, D. M. Huang, Y. C. Chen and C. Y. Mou, Chem. Mater., 2005, 17, 4570–4573 CrossRef CAS.
- T. H. Chung, S. H. Wu, M. Yao, C. W. Lu, Y. S. Lin, Y. Hung, C. Y. Mou, Y. C. Chen and D. M. Huang, Biomaterials, 2007, 28, 2959–2966 CrossRef CAS.
- A. G. Tkachenko, H. Xie, Y. L. Liu, D. Coleman, J. Ryan, W. R. Glomm, M. K. Shipton, S. Franzen and D. L. Feldheim, Bioconjugate Chem., 2004, 15, 482–490 CrossRef CAS.
- H. Takahashi, T. Niidome, T. Kawano, S. Yamada and Y. Niidome, J. Nanopart. Res., 2008, 10, 221–228 CrossRef CAS.
- T. B. Huff, M. N. Hansen, Y. Zhao, J. X. Cheng and A. Wei, Langmuir, 2007, 23, 1596–1599 CrossRef CAS.
- V. S. Kalambur, E. K. Longmire and J. C. Bischof, Langmuir, 2007, 23, 12329–12336 CrossRef CAS.
- T. Mosmann, J. Immunol. Methods, 1983, 65, 55–63 CrossRef CAS.
- C. M. Sayes, K. L. Reed and D. B. Warheit, Toxicol. Sci., 2007, 97, 163–180 CrossRef CAS.
- C. M. Sayes, J. D. Fortner, W. Guo, D. Lyon, A. M. Boyd, K. D. Ausman, Y. J. Tao, B. Sitharaman, L. J. Wilson, J. B. Hughes, J. L. West and V. L. Colvin, Nano Lett., 2004, 4, 1881–1887 CrossRef CAS.
- A. R. Simioni, F. L. Primo, M. M. A. Rodrigues, Z. G. M. Lacava, P. C. Morais and A. C. Tedesco, IEEE Trans. Magn., 2007, 43, 2459–2461 CrossRef CAS.
- G. N. Guo, W. Liu, J. G. Liang, Z. K. He, H. B. Xu and X. L. Yang, Mater. Lett., 2007, 61, 1641–1644 CrossRef CAS.
- A. J. Wagner, C. A. Bleckmann, R. C. Murdock, A. M. Schrand, J. J. Schlager and S. M. Hussain, J. Phys. Chem. B, 2007, 111, 7353–7359 CrossRef CAS.
- H. Dumortier, S. Lacotte, G. Pastorin, R. Marega, W. Wu, D. Bonifazi, J. P. Briand, M. Prato, S. Muller and A. Bianco, Nano Lett., 2006, 6, 1522–1528 CrossRef CAS.
- A. M. Derfus, W. C. W. Chan and S. N. Bhatia, Nano Lett., 2004, 4, 11–18 CrossRef CAS.
- G. Jia, H. F. Wang, L. Yan, X. Wang, R. J. Pei, T. Yan, Y. L. Zhao and X. B. Guo, Environ. Sci. Technol., 2005, 39, 1378–1383 CrossRef CAS.
- T. J. Brunner, P. Wick, P. Manser, P. Spohn, R. N. Grass, L. K. Limbach, A. Bruinink and W. J. Stark, Environ. Sci. Technol., 2006, 40, 4374–4381 CrossRef CAS.
- N. J. Marshall, C. J. Goodwin and S. J. Holt, Growth Regul., 1995, 5, 69–84 Search PubMed.
- T. Bernas and J. W. Dobrucki, Arch. Biochem. Biophys., 2000, 380, 108–116 CrossRef CAS.
- T. Bernas and J. Dobrucki, Cytometry, 2002, 47, 236–242 CrossRef CAS.
- R. J. Gonzalez and J. B. Tarloff, Toxicol. in Vitro, 2001, 15, 257–259 CrossRef CAS.
- S. A. B. Jabbar, P. R. Twentyman and J. V. Watson, Br. J. Cancer, 1989, 60, 523–528 CAS.
- B. L. Molinari, D. R. Tasat, M. A. Palmieri, S. E. O'Connor and R. L. Cabrini, Anal. Quant. Cytol. Histol., 2003, 25, 254–262 Search PubMed.
- D. Funk, H. H. Schrenk and E. Frei, BioTechniques, 2007, 43, 178–186 CrossRef CAS.
- K. Abe and H. Saito, Neurosci. Res., 1999, 35, 165–174 CrossRef CAS.
- S. Ahmad, A. Ahmad, K. B. Schneider and C. W. White, Int. J. Toxicol., 2006, 25, 17–23 CrossRef CAS.
- M. Natarajan, S. Mohan, B. R. Martinez, M. L. Meltz and T. S. Herman, Cancer Detect. Prev., 2000, 24, 405–414 Search PubMed.
- L. Belyanskaya, P. Manser, P. Spohn, A. Bruinink and P. Wick, Carbon, 2007, 45, 2643–2648 CrossRef CAS.
- K. Pulskamp, S. Diabate and H. F. Krug, Toxicol. Lett., 2007, 168, 58–74 CrossRef CAS.
- A. Casey, E. Herzog, M. Davoren, F. M. Lyng, H. J. Byrne and G. Chambers, Carbon, 2007, 45, 1425–1432 CrossRef CAS.
- T. Laaksonen, H. Santos, H. Vihola, J. Salonen, J. Riikonen, T. Heikkila, L. Peltonen, N. Kurnar, D. Y. Murzin, V. P. Lehto and J. Hirvonent, Chem. Res. Toxicol., 2007, 20, 1913–1918 CrossRef CAS.
- S. P. Low, K. A. Williams, L. T. Canham and N. H. Voelcker, Biomaterials, 2006, 27, 4538–4546 CrossRef CAS.
- N. A. Monteiro-Riviere and A. O. Inman, Carbon, 2006, 44, 1070–1078 CrossRef CAS.
- G. Punshon, D. S. Vara, K. M. Sales, A. G. Kidane, H. J. Salacinski and A. M. Seifalian, Biomaterials, 2005, 26, 6271–6279 CrossRef CAS.
- S. M. Hussain and J. M. Frazier, Toxicol. Sci., 2002, 69, 424–432 CrossRef CAS.
- S. N. Orlov, D. V. Pchejetski, S. D. Sarkissian, V. Adarichev, S. Taurin, A. V. Pshezhetsky, J. Tremblay, G. V. Maximov, D. deBlois, M. R. Bennett and P. Hamet, Apoptosis, 2003, 8, 199–208 Search PubMed.
- T. T. Puck and P. I. Marcus, J. Exp. Med., 1956, 103, 653–666 CrossRef CAS.
- N. A. Franken, H. M. Rodermond, J. Stap, J. Haveman and C. van Bree, Nat. Protoc., 2006, 1, 2315–2319 Search PubMed.
- C. Zhang, B. Wangler, B. Morgenstern, H. Zentgraf, M. Eisenhut, H. Untenecker, R. Kruger, R. Huss, C. Seliger, W. Semmler and F. Kiessling, Langmuir, 2007, 23, 1427–1434 CrossRef CAS.
- N. A. Monteiro-Riviere, R. J. Nemanich, A. O. Inman, Y. Y. Y. Wang and J. E. Riviere, Toxicol. Lett., 2005, 155, 377–384 CrossRef CAS.
- W. F. Vevers and A. N. Jha, Ecotoxicology, 2008, 17, 410–420 CrossRef CAS.
- I. Nicoletti, G. Migliorati, M. C. Pagliacci, F. Grignani and C. Riccardi, J. Immunol. Methods, 1991, 139, 271–279 CrossRef CAS.
- K. J. Lee, P. D. Nallathamby, L. M. Browning, C. J. Osgood and X. H. N. Xu, ACS Nano, 2007, 1, 133–143 CrossRef CAS.
- H. M. Evans and W. Schulemann, Science (Washington, DC, U.S.), 1914, 39, 443–454 Search PubMed.
- E. Borenfreund and J. A. Puerner, Toxicol. Lett., 1985, 24, 119–124 CrossRef CAS.
- Y. H. Jin, S. Kannan, M. Wu and J. X. J. Zhao, Chem. Res. Toxicol., 2007, 20, 1126–1133 CrossRef CAS.
- T. Decker and M. L. Lohmann-Matthes, J. Immunol. Methods, 1988, 115, 61–69 CrossRef CAS.
- C. Korzeniewski and D. M. Callewaert, J. Immunol. Methods, 1983, 64, 313–320 CrossRef CAS.
- L. R. Hirsch, R. J. Stafford, J. A. Bankson, S. R. Sershen, B. Rivera, R. E. Price, J. D. Hazle, N. J. Halas and J. L. West, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 13549–13554 CrossRef CAS.
- C. L. Nehl, N. K. Grady, G. P. Goodrich, F. Tam, N. J. Halas and J. H. Hafner, Nano Lett., 2004, 4, 2355–2359 CrossRef CAS.
- E. S. Kaneshiro, M. A. Wyder, Y. P. Wu and M. T. Cushion, J. Microbiol. Methods, 1993, 17, 1–16 CrossRef CAS.
- M. Bottini, S. Bruckner, K. Nika, N. Bottini, S. Bellucci, A. Magrini, A. Bergamaschi and T. Mustelin, Toxicol. Lett., 2006, 160, 121–126 CrossRef CAS.
- D. X. Cui, F. R. Tian, C. S. Ozkan, M. Wang and H. J. Gao, Toxicol. Lett., 2005, 155, 73–85 CrossRef CAS.
- A. Omidkhoda, H. Mozdarani, A. Movasaghpoor and A. A. P. Fatholah, Toxicol. in Vitro, 2007, 21, 1191–1196 CrossRef CAS.
- K. C. Bhol and P. J. Schechter, Br. J. Dermatol., 2005, 152, 1235–1242 CrossRef CAS.
- Y. Mo and L. Y. Lim, J. Controlled Release, 2005, 108, 244–262 CrossRef CAS.
- M. van Engeland, L. J. W. Nieland, F. C. S. Ramaekers, B. Schutte and C. P. M. Reutelingsperger, Cytometry, 1998, 31, 1–9 CrossRef CAS.
- M. J. Arends, R. G. Morris and A. H. Wyllie, Am. J. Pathol., 1990, 136, 593–608 Search PubMed.
- S. Nagata, H. Nagase, K. Kawane, N. Mukae and H. Fukuyama, Cell Death Differ., 2003, 10, 108–116 CrossRef CAS.
- N. P. Singh, M. T. McCoy, R. R. Tice and E. L. Schneider, Exp. Cell Res., 1988, 175, 184–191 CrossRef CAS.
- N. R. Jacobsen, G. Pojana, P. White, P. Moller, C. A. Cohn, K. S. Korsholm, U. Vogel, A. Marcomini, S. Loft and H. Wallin, Environ. Mol. Mutagen., 2008, 49, 476–487 CrossRef CAS.
- C. A. Barnes, A. Elsaesser, J. Arkusz, A. Smok, J. Palus, A. Lesniak, A. Salvati, J. P. Hanrahan, W. H. de Jong, E. Dziubaltowska, M. Stepnik, K. Rydzynski, G. McKerr, I. Lynch, K. A. Dawson and C. V. Howard, Nano Lett., 2008, 8, 3069–3074 CrossRef CAS.
- H. L. Karlsson, P. Cronholm, J. Gustafsson and L. Moller, Chem. Res. Toxicol., 2008, 21, 1726–1732 CrossRef CAS.
- B. M. K. Y. J. L. H. W. C. Su Jin Kang, Environ. Mol. Mutagen., 2008, 49, 399–405 CrossRef CAS.
- P. Mroz, A. Pawlak, M. Satti, H. Lee, T. Wharton, H. Gali, T. Sarna and M. R. Hamblin, Free Radical Biol. Med., 2007, 43, 711–719 CrossRef CAS.
- R. Colognato, A. Bonelli, J. Ponti, M. Farina, E. Bergamaschi, E. Sabbioni and L. Migliore, Mutagenesis, 2008, 23, 377–382 CrossRef CAS.
- E. R. Kisin, A. R. Murray, M. J. Keane, X. C. Shi, D. Schwegler-Berry, O. Gorelik, S. Arepalli, V. Castranova, W. E. Wallace, V. E. Kagan and A. A. Shvedova, J. Toxicol. Environ. Health, Part A, 2007, 70, 2071–2079 Search PubMed.
- Y. Gavrieli, Y. Sherman and S. A. Bensasson, J. Cell Biol., 1992, 119, 493–501 CrossRef.
- X. Li and Z. Darzynkiewicz, Cell Proliferation, 1995, 28, 571–579 CrossRef CAS.
- S. Magder, Crit. Care, 2006, 10(1), 208 CrossRef.
- J. M. Mates, Toxicology, 2000, 153, 83–104 CrossRef CAS.
- A. G. Fantel, Teratology, 1996, 53, 196–217 CrossRef CAS.
- C. M. Sayes, A. M. Gobin, K. D. Ausman, J. Mendez, J. L. West and V. L. Colvin, Biomaterials, 2005, 26, 7587–7595 CrossRef CAS.
- O. Choi and Z. Q. Hu, Environ. Sci. Technol., 2008, 42, 4583–4588 CrossRef CAS.
- T. C. Long, N. Saleh, R. D. Tilton, G. V. Lowry and B. Veronesi, Environ. Sci. Technol., 2006, 40, 4346–4352 CrossRef CAS.
- A. M. Schrand, L. K. Braydich-Stolle, J. J. Schlager, L. M. Dai and S. M. Hussain, Nanotechnology, 2008, 19, 235104 CrossRef.
- S. J. Clarke, C. A. Hollmann, Z. J. Zhang, D. Suffern, S. E. Bradforth, N. M. Dimitrijevic, W. G. Minarik and J. L. Nadeau, Nat. Mater., 2006, 5, 409–417 CrossRef CAS.
- W. S. Lin, Y. W. Huang, X. D. Zhou and Y. F. Ma, Toxicol. Appl. Pharmacol., 2006, 217, 252–259 CrossRef CAS.
- A. Isakovic, Z. Markovic, B. Todorovic-Markovic, N. Nikolic, S. Vranjes-Djuric, M. Mirkovic, M. Dramicanin, L. Harhaji, N. Raicevic, Z. Nikolic and V. Trajkovic, Toxicol. Sci., 2006, 91, 173–183 CrossRef CAS.
- L. Y. Zang, B. R. Misra, F. vanKuijk and H. P. Misra, Biochem. Mol. Biol. Int., 1995, 37, 1187–1195 CAS.
- A. Gomes, E. Fernandes and J. Lima, J. Biochem. Biophys. Methods, 2005, 65, 45–80 CrossRef CAS.
- B. Halliwell and M. Whiteman, Br. J. Pharmacol., 2004, 142, 231–255 CrossRef CAS.
- D. A. Bass, J. W. Parce, L. R. Dechatelet, P. Szejda, M. C. Seeds and M. Thomas, J. Immunol., 1983, 130, 1910–1917 CAS.
- C. Hanley, J. Layne, A. Punnoose, K. M. Reddy, I. Coombs, A. Coombs, K. Feris and D. Wingett, Nanotechnology, 2008, 19, 295103 CrossRef.
- W. S. Lin, Y. W. Huang, X. D. Zhou and Y. F. Ma, Int. J. Toxicol., 2006, 25, 451–457 CrossRef CAS.
- Y. K. Mishra, S. Mohapatra, D. K. Avasthi, D. Kabiraj, N. P. Lalla, J. C. Pivin, H. Sharma, R. Kar and N. Singh, Nanotechnology, 2007, 18, 345606 CrossRef.
- T. C. Long, J. Tajuba, P. Sama, N. Saleh, C. Swartz, J. Parker, S. Hester, G. V. Lowry and B. Veronesi, Environ. Health Perspect., 2007, 115, 1631–1637 CAS.
- A. Lawrence, C. M. Jones, P. Wardman and M. J. Burkitt, J. Biol. Chem., 2003, 278, 29410–29419 CrossRef CAS.
- C. P. Wan, E. Myung and B. H. S. Lau, J. Immunol. Methods, 1993, 159, 131–138 CrossRef CAS.
- S. L. Hempel, G. R. Buettner, Y. Q. O'Malley, D. A. Wessels and D. M. Flaherty, Free Radical Biol. Med., 1999, 27, 146–159 CrossRef CAS.
- K. M. Robinson, M. S. Janes, M. Pehar, J. S. Monette, M. F. Ross, T. M. Hagen, M. P. Murphy and J. S. Beckman, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 15038–15043 CrossRef CAS.
- K. M. Robinson, M. S. Janes and J. S. Beckman, Nat. Protoc., 2008, 3, 941–947 Search PubMed.
- E. H. W. Pap, G. P. C. Drummen, V. J. Winter, T. W. A. Kooij, P. Rijken, K. W. A. Wirtz, J. A. F. Op den Kamp, W. J. Hage and J. A. Post, FEBS Lett., 1999, 453, 278–282 CrossRef CAS.
- J. M. Worle-Knirsch, K. Kern, C. Schleh, C. Adelhelm, C. Feldmann and H. F. Krug, Environ. Sci. Technol., 2007, 41, 331–336 CrossRef.
- C. M. Sayes, A. A. Marchione, K. L. Reed and D. B. Warheit, Nano Lett., 2007, 7, 2399–2406 CrossRef CAS.
- A. A. Shvedova, V. Castranova, E. R. Kisin, D. Schwegler-Berry, A. R. Murray, V. Z. Gandelsman, A. Maynard and P. Baron, J. Toxicol. Environ. Health, Part A, 2003, 66, 1909–1926 Search PubMed.
- V. E. Kagan, Y. Y. Tyurina, V. A. Tyurin, N. V. Konduru, A. I. Potapovich, A. N. Osipov, E. R. Kisin, D. Schwegler-Berry, R. Mercer, V. Castranova and A. A. Shvedova, Toxicol. Lett., 2006, 165, 88–100 CrossRef CAS.
- S. M. Hussain, A. K. Javorina, A. M. Schrand, H. M. Duhart, S. F. Ali and J. J. Schlager, Toxicol. Sci., 2006, 92, 456–463 CrossRef CAS.
- J. Lakritz, C. G. Plopper and A. R. Buckpitt, Anal. Biochem., 1997, 247, 63–68 CrossRef CAS.
- C. S. Sharma, S. Sarkar, A. Periyakaruppan, J. Barr, K. Wise, R. Thomas, B. L. Wilson and G. T. Ramesh, J. Nanosci. Nanotechnol., 2007, 7, 2466–2472 CrossRef CAS.
- I. Rahman, A. Kode and S. K. Biswas, Nat. Protoc., 2006, 1, 3159–3165 Search PubMed.
- R. Brigelius, C. Muckel, T. P. Akerboom and H. Sies, Biochem. Pharmacol., 1983, 32, 2529–2534 CrossRef CAS.
- H. Güntherberg and J. Rost, Anal. Biochem., 1966, 15, 205–210 CrossRef CAS.
- Y. Sun, L. W. Oberley and Y. Li, Clin. Chem. (Washington, D. C.), 1988, 34, 497–500 CAS.
- T. T. Zhang, J. L. Stilwell, D. Gerion, L. H. Ding, O. Elboudwarej, P. A. Cooke, J. W. Gray, A. P. Alivisatos and F. F. Chen, Nano Lett., 2006, 6, 800–808 CrossRef CAS.
- K. A. Giuliano, R. L. DeBiasio, R. T. Dunlay, A. Gough, J. M. Volosky, J. Zock, G. N. Pavlakis and D. L. Taylor, J. Biomol. Screening, 1997, 2, 249–259 CrossRef CAS.
- K. A. Giuliano, J. R. Haskins and D. L. Taylor, Assay Drug Dev. Technol., 2003, 1, 565–577 CrossRef CAS.
- E. Jan, S. J. Byrne, M. Cuddihy, A. M. Davies, Y. Volkov, Y. K. Gun'ko and N. A. Kotov, ACS Nano, 2008, 2, 928–938 CrossRef CAS.
- B. J. Marquis, A. D. McFarland, K. L. Braun and C. L. Haynes, Anal. Chem., 2008, 80, 3431–3437 CrossRef CAS.
- R. Kopelman, Y.-E. Lee Koo, M. Philbert, B. A. Moffat, G. Ramachandra Reddy, P. McConville, D. E. Hall, T. L. Chenevert, M. S. Bhojani, S. M. Buck, A. Rehemtulla and B. D. Ross, J. Magn. Magn. Mater., 2005, 293, 404–410 CrossRef CAS.
- C. M. Ensor, F. W. Holtsberg, J. S. Bomalaski and M. A. Clark, Cancer Res., 2002, 62, 5443–5450 CAS.
- D. Shenoy, S. Little, R. Langer and M. Amiji, Pharmacol. Res., 2005, 22, 2107–2114 CrossRef CAS.
- M. T. Zhu, W. Y. Feng, B. Wang, T. C. Wang, Y. Q. Gu, M. Wang, Y. Wang, H. Ouyang, Y. L. Zhao and Z. F. Chai, Toxicology, 2008, 247, 102–111 CrossRef CAS.
- S. C. Kim, D. W. Kim, Y. H. Shim, J. S. Bang, H. S. Oh, S. W. Kim and M. H. Seo, J. Controlled Release, 2001, 72, 191–202 CrossRef CAS.
- G. L. Baker, A. Gupta, M. L. Clark, B. R. Valenzuela, L. M. Staska, S. J. Harbo, J. T. Pierce and J. A. Dill, Toxicol. Sci., 2008, 101, 122–131 CAS.
- F. Alexis, E. Pridgen, L. K. Molnar and O. C. Farokhzad, Mol. Pharmacol., 2008, 5, 505–515 CrossRef CAS.
- D. E. Owens and N. A. Peppas, Int. J. Pharm., 2006, 307, 93–102 CrossRef CAS.
- X. K. Sun, R. Rossin, J. L. Turner, M. L. Becker, M. J. Joralemon, M. J. Welch and K. L. Wooley, Biomacromolecules, 2005, 6, 2541–2554 CrossRef CAS.
- T. K. Jain, M. K. Reddy, M. A. Morales, D. L. Leslie-Pelecky and V. Labhasetwar, Mol. Pharmacol., 2008, 5, 316–327 CrossRef CAS.
- X. Gao, Y. Cui, R. M. Levenson, L. W. K. Chung and S. Nie, Nat. Biotechnol., 2004, 22, 969–976 CrossRef CAS.
- H. C. Fischer, L. C. Liu, K. S. Pang and W. C. W. Chan, Adv. Funct. Mater., 2006, 16, 1299–1305 CrossRef CAS.
- M. Q. Chu, Q. Wu, J. X. Wang, S. K. Hou, Y. Miao, J. L. Peng and Y. Sun, Nanotechnology, 2007, 18, 455103 CrossRef.
- Z. Chen, H. Meng, G. Xing, C. Chen, Y. Zhao, G. Jia, T. Wang, H. Yuan, C. Ye, F. Zhao, Z. Chai, C. Zhu, X. Fang, B. Ma and L. Wan, Toxicol. Lett., 2006, 163, 109–120 CrossRef CAS.
- B. Wang, W. Y. Feng, T. C. Wang, J. Guang, M. Wang, J. W. Shi, F. Zhang, Y. L. Zhao and Z. F. Chai, Toxicol. Lett., 2006, 161, 115–123 CrossRef CAS.
- E. I. Altinoglu, T. J. Russin, J. M. Kaiser, B. M. Barth, P. C. Eklund, M. Kester and J. H. Adair, ACS Nano, 2008, 2, 2075–2084 CrossRef CAS.
- Y. P. Li, Y. Y. Pei, X. Y. Zhang, Z. H. Gu, Z. H. Zhou, W. F. Yuan, J. J. Zhou, J. H. Zhu and X. J. Gao, J. Controlled Release, 2001, 71, 203–211 CrossRef CAS.
- X. L. Huang, B. Zhang, L. Ren, S. F. Ye, L. P. Sun, Q. Q. Zhang, M. C. Tan and G. M. Chow, J. Mater. Sci.: Mater. Med., 2008, 19, 2581–2588 CrossRef CAS.
- P. Lin, J. W. Chen, L. W. Chang, J. P. Wu, L. Redding, H. Chang, T. K. Yeh, C. S. Yang, M. H. Tsai, H. J. Wang, Y. C. Kuo and R. S. H. Yang, Environ. Sci. Technol., 2008, 42, 6264–6270 CrossRef CAS.
- L. C. Lin, K. Y. Yang, Y. F. Chen, S. C. Wang and T. H. Tsai, J. Chromatogr., A, 2005, 1073, 285–289 CrossRef CAS.
- R. H. Lei, C. Q. Wu, B. H. Yang, H. Z. Ma, C. Shi, Q. J. Wang, Q. X. Wang, Y. Yuan and M. Y. Liao, Toxicol. Appl. Pharmacol., 2008, 232, 292–301 CrossRef CAS.
- L. J. Mortensen, G. Oberdorster, A. P. Pentland and L. A. DeLouise, Nano Lett., 2008, 8, 2779–2787 CrossRef CAS.
- A. Flesken-Nikitin, I. Toshkov, J. Naskar, K. M. Tyner, R. M. Williams, W. R. Zipfel, E. P. Giannelis and A. Y. Nikitin, Toxicol. Pathol., 2007, 35, 804–810 CrossRef CAS.
- J. Muller, F. Huaux, A. Fonseca, J. B. Nagy, N. Moreau, M. Delos, E. Raymundo-Pinero, F. Beguin, M. Kirsch-Volders, I. Fenoglio, B. Fubini and D. Lison, Chem. Res. Toxicol., 2008, 21, 1698–1705 CrossRef CAS.
- Z. D. Sandlin, M. Shou, J. G. Shackman and R. T. Kennedy, Anal. Chem., 2005, 77, 7702–7708 CrossRef CAS.
- N. A. Cellar, S. T. Burns, J. C. Meiners, H. Chen and R. T. Kennedy, Anal. Chem., 2005, 77, 7067–7073 CrossRef CAS.
- B. L. Hogan, S. M. Lunte, J. F. Stobaugh and C. E. Lunte, Anal. Chem., 1994, 66, 596–602 CrossRef CAS.
- S. Y. Zhou, H. Zuo, J. F. Stobaugh, C. E. Lunte and S. M. Lunte, Anal. Chem., 1995, 67, 594–599 CrossRef CAS.
- J. F. Cheer, B. J. Aragona, M. Heien, A. T. Seipel, R. M. Carelli and R. M. Wightman, Neuron, 2007, 54, 237–244 CrossRef CAS.
- B. J. Aragona, N. A. Cleaveland, G. D. Stuber, J. J. Day, R. M. Carelli and R. M. Wightman, J. Neurosci., 2008, 28, 8821–8831 CrossRef CAS.
- A. G. Ewing, J. C. Bigelow and R. M. Wightman, Science, 1983, 221, 169–171 CAS.
- Y. T. Wu, Y. F. Chen, Y. J. Hsieh, I. Jaw, M. S. Shiao and T. H. Tsai, Int. J. Pharm., 2006, 326, 25–31 CrossRef CAS.
- X. L. Kong, L. C. L. Huang, C. M. Hsu, W. H. Chen, C. C. Han and H. C. Chang, Anal. Chem., 2005, 77, 259–265 CrossRef CAS.
- S. Y. Chang, N. Y. Zheng, C.-S. Chen, C.-D. Chen, Y.-Y. Chen and C. R. C. Wang, J. Am. Soc. Mass Spectrom., 2007, 18, 910–918 CrossRef CAS.
- D. A. Stuart, J. M. Yuen, N. Shah, O. Lyandres, C. R. Yonzon, M. R. Glucksberg, J. T. Walsh and R. P. Van Duyne, Anal. Chem., 2006, 78, 7211–7215 CrossRef CAS.
- J. Kneipp, H. Kneipp, M. McLaughlin, D. Brown and K. Kneipp, Nano Lett., 2006, 6, 2225–2231 CrossRef CAS.
- J. Kneipp, H. Kneipp, B. Wittig and K. Kneipp, Nano Lett., 2007, 7, 2819–2823 CrossRef CAS.
- C. Loo, A. Lowery, N. Halas, J. West and R. Drezek, Nano Lett., 2005, 5, 709–711 CrossRef CAS.
- T. Lühmann, M. Rimann, A. G. Bittermann and H. Hall, Bioconjugate Chem., 2008, 19, 1907–1916 CrossRef CAS.
- C. Muhlfeld, B. Rothen-Rutishauser, D. Vanhecke, F. Blank, P. Gehr and M. Ochs, Part. Fibre Toxicol., 2007, 4, 1–17 CrossRef.
- S. Lee, S. Choi Joon and H. Kang Seong, J. Nanosci. Nanotechnol., 2007, 7, 3689–3694 CrossRef CAS.
- G. Oberdorster, E. Oberdorster and J. Oberdorster, Environ. Health Perspect., 2007, 115, A290.
- G. Oberdorster, E. Oberdorster and J. Oberdorster, Environ. Health Perspect., 2005, 113, 823–839 CAS.
- W. G. Kreyling, M. Semmler-Behnke and W. Moeller, J. Nanopart. Res., 2006, 8, 543–562 CrossRef CAS.
- P. J. A. Borm and R. P. F. Schins, Drugs Pharm. Sci., 2006, 159, 161–197 Search PubMed.
- K. Donaldson, R. Aitken, L. Tran, V. Stone, R. Duffin, G. Forrest and A. Alexander, Toxicol. Sci., 2006, 92, 5–22 CrossRef CAS.
- K. Donaldson, L. Tran, L. A. Jimenez, R. Duffin, D. E. Newby, N. Mills, W. MacNee and V. Stone, Part. Fibre Toxicol., 2005, 2, 1–14 CrossRef.
| This journal is © The Royal Society of Chemistry 2009 |
