DNA-templated CMV viral capsidproteins assemble into nanotubes†
Yun
Xu‡
a,
Jian
Ye‡
b,
Huajie
Liu
a,
Enjun
Cheng
a,
Yang
Yang
a,
Wenxing
Wang
a,
Manchun
Zhao
a,
Dejian
Zhou
c,
Dongsheng
Liu
*a and
Rongxiang
Fang
b
aNational Centre for NanoScience & Technology, No. 11, Beiyitiao, Zhongguancun, Beijing 100080, P. R. China. E-mail: liuds@nanoctr.cn; Fax: +86-10-62656765; Tel: +86-10-82545589
bState Key Laboratory of Plant Genomics, Institute of Microbiology, Chinese Academy of Sciences, Beijing 100101, P. R. China
cSchool of Chemistry and Astbury Centre for Structural Molecular Biology, University of Leeds, Leeds, UK LS2 9JT
First published on 21st November 2007
Abstract
This communication describes the in vitro assembly of genetically recombinant Cucumber Mosaic Virus (CMV) viral capsidproteins (CPs) into biological nanotubes, several micrometres long yet with a diameter of only ∼17 nm, triggered by double-stranded DNAs of different lengths.
Viral capsid proteins (CPs) have multiple functions and play an important role in the life cycle of a virus, such as self-assembly into virus-like particles and encapsulation of a viral genome, selective recognition of target cells through binding to membrane receptors, etc. In addition to its significance in viral life, the desirable packaging and self-assembly properties, the nanometre size scale and easy functionalization characteristics have made capsids ideal building materials for nanotechnology.1,2 For example, the CP capsid offers a constrained and monodispersed system that can be exploited as a container for gold and other nanoparticles, and provides protection for the encapsidated cargo.3,4 The regular motifs of tubular or quasi-spherical capsids have been used for the templated synthesis of nanomaterials ranging from magnetic particles to nanowires for electronic applications.5,6 Recently, Zlotnick found that, as well as being able to self-assemble into hollow particles, CPs of Cowpea chlorotic mottle virus (CCMV), one of the most intensively studied spherical plant viruses, could assemble into tubular nanostructures templated by heterogeneous DNA longer than 500 base pairs (bps) in vitro.7 This finding pilots a new way to prepare nanotubes with biomolecules. Meanwhile, many aspects of the assembly process should be improved in order to make it applicable. In this communication, we describe our discovery that protein nanotubes with lengths of over several micrometres can be assembled with engineered Cucumber Mosaic Virus (CMV) capsid protein templated by double-stranded DNA as short as 50 bp, which has long been thought impossible without RNA either in vivo or in vitro.8
The protein used in our assembly is the CP of CMV, a typical member of the genus Cucumovirus (family Bromoviridae), which has the broadest host range of any known virus, infecting more than 1000 species of plants and causing huge economic losses for many crops worldwide.8,9 This T = 3 virus is best represented as a truncated icosahedron with three copies of capsid protein in each icosahedral asymmetric unit. In the previous report, the CCMV CP was purified from viron particles extracted from virus infected plant materials, which are virus replication dependent, susceptible to protease degradation and unable to be produced on a large scale.7 As with the reported CCMV system, expression of the CMV CPs by the widely used traditional E. coli expression system can only give insoluble inclusion bodies or a very low quantity of soluble proteins . To overcome this limitation, we have developed a novel recombination procedure to produce a mass of pure soluble CMV CPs: firstly, the CP gene was cloned into the expression vector pET11a (Novagen, USA); after expression, the inclusion bodies were separated and purified, then solubilized in a 2 M urea solution by denaturation; the denatured protein solution was then applied to a refolding procedure where the soluble denatured CP was allowed to refold at a relatively low concentration (see ESI for detailed experimental procedures and conditions† ). Before being applied to the assembly process, the refolded CP was concentrated through an ultrafiltration system to obtain a final concentration of 50 µg ml−1 in Tris buffer (20 mM Tris-HCl, 150 mM NaCl, pH 7.5).
Fig. 1 shows the assembly process schematically. The detailed experimental procedures are described in the ESI.† Briefly, 0.1 µM 500 bp Polymerase Chain Reaction (PCR) amplified double-stranded DNA (see ESI for details† ) was mixed with the purified CP (50 µg ml−1) in Tris buffer, where the substrates were vaired from 1 to 20 DNA bp per CP. The mixture was then incubated at room temperature for 20 min to obtain the assembled structure.10
 | ||
| Fig. 1 Schematic of the formation of biological nanotubes by DNA directed assembly of CP. | ||
The assembled tubular structures were firstly investigated by transmission electron microscopy (TEM ). The products were applied onto copper grids coated with amorphous carbon film and then negatively stained with 2% uranyl acetate.11 As shown in Fig. 2, uniform tubular structures were regularly observed. Most of the tubes were longer than a micrometre (Fig. 2B) with a narrow diameter distribution around 17 nm. This suggests that these DNA templated self-assembled tubular structures are highly uniform. A few much bigger tubular structures were also observed in some of the TEM images, which may result from the formation of bundles of these individual tubular structures. In contrast, no tubular structures were observed in control experiments where either the DNA or the CP was absent under identical conditions (see ESI Fig. S2† ). This clearly demonstrates that both the DNA template and the CP are essential to the formation of the tubular structures observed here.
 | ||
| Fig. 2 (Upper) Representative TEM images of uranyl acetate stained tubular structures formed by CMV CP and non-specific 500 bp dsDNA. The scale bars are 50 nm. (Lower) Histograms show the wide variation in the length and the narrow variation in the width of tubes in micrographs. Lengths and widths were measured from 25 micrographs. | ||
To demonstrate that these tubular structures were formed by the CP templated by DNA, the following experiments were carried out: (1) circular dichroism (CD) spectroscopy was employed to study the interaction between the DNA and the CP. As shown in Fig. 3, the DNA used in the experiment had a CD spectrum which showed positive absorbance at 215 nm, 270 nm and negative absorbance at 200 nm, 220 nm (green line), which is a distinctive characteristic for a double-stranded structure.12 The purified and refolded CP had a CD spectrum with a positive absorbance at 191 nm and a negative absorbance at 210 nm (red line). The spectrum of the assembly mixture was compared with the simple addition of the DNA and CP spectra and the difference is shown in the insert of Fig. 3. The positive bands near 200 nm, 215 nm and a small negative band near 210 nm in the spectrum demonstrated that there is strong interaction between the DNA and the CP within the assembly. (2) Because the tubular structures are several micrometres long, so visible under an optical microscope, we could directly compare the optical and fluorescence images of the same structure. Here we used a fluorophore labeled DNA as the substrate to template the assembly of the CPs under identical conditions. The optical and corresponding fluorescence images clearly showed that the fluorophores (the DNA templates) were mostly included in tubular structures (see Fig. S1, ESI† ). This result further demonstrated that the tubular structures were indeed assembled from the CPs on the DNA template.
 | ||
| Fig. 3 CD spectra of dsDNA (green line), CMV CP (red line) and assembly products (blue line). The difference between the spectrum of the assembly mixture and CP spectra + DNA spectra is shown as an insert. | ||
To investigate whether the length of the DNA template is critical to the nanotube formation, we have explored the assembly of CP with shorter template DNAs. Unlike the reported CCMV–DNA system,7 where a relatively long DNA template of over 500 bps was required, our results showed that the CMV CP could assemble with 300 bp, 100 bp and even 50 bp DNAs and produced similar structures (Fig. 4A, B and C) under the same conditions. The tubular structures obtained with these shorter DNA templates had very similar diameters to those obtained with the longer 500 bp DNAs. How is the tubular structure formed? Assembly, like crystallization, is thought to proceed through separate nucleation and growth events. Thus, the tube assembly may start from a nucleation center involving several DNA and CP molecules. In addition to the helical geometry model supported by a scaffold of staggered DNA strands,7 the CP–DNA complex may alternatively result from stacks of rings (Fig. 1).
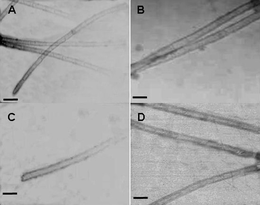 | ||
| Fig. 4 TEM images of the tubular structures formed by the CMV CP with non-specific dsDNA of different lengths. (A) 300 bps DNA, (B) 100 bps DNA, (C) 50 bps DNA and (D) SARS-CoV N gene fragments (about 500 bps). Scale bars are all 50 nm. | ||
To further investigate the potential biomedical use of these nanostructures, we also used a DNA fragment of SARS-CoV (severe acute respiratory syndrome-associated coronavirus, about 500 bp long) as the template, which is normally used to express small interfering RNA (siRNA) for RNAi-based therapy.13 Similar tubular structures were obtained (Fig. 4D). The CMV CP contains a polyarginine domain,14 which is part of the HIV-1 Tat protein transduction domain and has been demonstrated to play an important role in its cell uptake.15,16 Thus the DNA templated CP assembly developed here could not only benefit the fabrication of novel biological nanostructures but also possesses great potential for the development of carriers useful for gene delivery, as well as DNA or RNA aptamer delivery.
In summary, we have developed a new method to scale up the production of pure CMV CPs. We have also demonstrated that this protein can assemble into tubular structures in the presence of heterogeneous double-stranded DNA templates of various lengths down to 50 bps in vitro. The resulting nanotubes were several micrometres long yet with a diameter of only around 17 nm, comparable to amyloid fibres. CD spectroscopy and fluorescent microscopy confirmed that these nanotubes were formed from the assembly of the CPs on the DNA template. These biologically compatible nanotubes (made up entirely from proteins and DNAs) may have potential as gene/drug carriers. By combining protein engineering with DNA template design, this development will benefit the design and assembly of more complicated, multi-functional bio-nanomaterials.
The authors thank Prof. D. Han, Prof. Y. Zheng and Ms Y. Xie, NCNST, and Mr Y. Sun, ICCAS, for their assistance in TEM and fluorescent imaging experiments. The authors also thank Prof. D. Kelnerman, University of Cambridge, for his constructive suggestions. This work was supported by grants from the Science 100 Program of CAS, the National Natural Science Foundation of China under grant No.20573027 and MOST under grant No. 2007CB935902.
Notes and references
- D. Trevor and M. Young, Nature, 1998, 393, 152 CrossRef CAS.
- C. Chen, E. S. Kwak, B. Stein, C. C. Kao and B. Dragnea, J. Nanosci. Nanotechnol., 2005, 5, 2029 CrossRef CAS.
- L. N. Loo, R. H. Guenther, V. R. Basnayake, S. A. Lommel and S. Franzen, J. Am. Chem. Soc., 2006, 128, 4502 CrossRef CAS.
- J. D. Lewis, G. Destito, A. Zijlstra, M. J. Gonzalez, J. P. Quigley, M. Manchester and H. Stuhlmann, Nat. Med., 2006, 12, 354 CrossRef CAS.
- S. Meunier, E. Strable and M. G. Finn, Science, 2004, 303, 213 CrossRef CAS.
- R. J. Tseng, C. G. Tsai, L. P. Ma, J. Y. Ouyang, C. S. Ozkan and Y. Yang, Nat. Nanotechnol., 2006, 1, 72 Search PubMed.
- S. Mukherjee, C. M. Pfeifer, J. M. Johnson and A. Zlotnick, J. Am. Chem. Soc., 2006, 128, 2538 CrossRef CAS.
- W. R. Wikoff, C. J. Tsai, G. J. Wang, T. S. Baker and J. E. Johnson, J. Virol., 1997, 232, 91 CrossRef CAS.
- T. J. Smith, E. Chase, T. Schmidt and K. L. Perry, J. Virol., 2000, 74, 7578 CrossRef CAS.
- This assembly process is highly reproducible. The experiments were repeated over 20 times with several different batches of CPs; all reliably produced protein nanotubes. We found that longer incubation time has little influence on the length distributions of the resulting nanotubes.
- K. Sugihara, P. A. Reichart, H. R. Gelderblom, H. D. Pohle, A. Langford and H. Reupke, Int. Conf. AIDS, 1990, 6, 20 Search PubMed.
- X. G. Sun, E. H. Cao, Y. J. He and J. F. Qin, J. Biomol. Struct. Dyn., 1999, 16, 863 CAS.
- B. J. Zheng, Y. Guan and Q. Tang, Antiviral Ther., 2004, 9, 365 Search PubMed.
- R. Tan and A. D. Frankel, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 5282 CrossRef CAS.
- A. Dragulescu-Andrasi, P. Zhou, G. He and D. H. Ly, Chem. Commun., 2005, 244 RSC.
- A. Dragulescu-Andrasi, S. Rapireddy, G. F. He, B. Bhattacharya, J. J. Hyldig-Nielsen, G. Zon and D. H. Ly, J. Am. Chem. Soc., 2006, 128, 16104 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Details of the experimental procedures, the optical and fluorescence images and gel electrophoresis graphs of CMV CPs. See DOI: 10.1039/b715299j |
| ‡ These two authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2008 |
