The RNA2–PNA2 hybrid i-motif—a novel RNA -based building block†
Saikat
Chakraborty
,
Souvik
Modi
and
Yamuna
Krishnan
*
National Centre for Biological Sciences, TIFR, GKVK, Bellary Road, Bangalore, 560065, India. E-mail: yamuna@ncbs.res.in; Fax: +91-80-23636662; Tel: +91-80-23666180
First published on 17th October 2007
Abstract
We report the formation of a hybrid RNA2–PNA2 i-motif comprised of two RNA and two PNA strands based on the sequence specific self assembly of RNA , with potential as a building block for structural RNA nanotechnology.
Structural DNA nanotechnology is a field where DNA is used to create robust and dynamic nanoscale architectures.1 We have been interested in exploring the potential of non-Watson–Crick base paired motifs in the rational construction of such architectures and towards this end developed many such non B-DNA building blocks.2RNA is now emerging as a versatile entity for the analogous RNA equivalent, structural RNA nanotechnology.3RNA is capable of much more structural variety than DNA and has the capability of forming several unusual motifs through a variety of tertiary interactions that are predominantly non-Watson–Crick type.4 Given this versatility, we sought to explore the predictable creation of RNA -based non-Watson–Crick building blocks for potential use as rational design elements in structural RNA technology.
C-rich sequences of DNA, RNA and their mimics are known to associate into a four-stranded structure, called the i-motif, that is held together by hemiprotonated C–C+ base paired duplexes that are intercalated in an antiparallel orientation.5 Given that RNA and PNA can form RNA4 and PNA4 i-motifs respectively, we wanted to see whether an equimolar mixture of C-rich RNA and PNA could form a hybrid i-motif.5 In this paper, we describe the formation of a unique population of a hybrid R2P2 i-motif from a binary mixture of C-rich RNA and PNA sequences.
We used C-rich PNA, P (Fig. 1) and C-rich RNA , R, that incorporated a T at the N-terminus and U at the 5′ end respectively to prevent higher order structure formation.2 Native polyacrylamide gel electrophoresis showed that a 1 : 1 mixture of R : P forms a hybrid complex comprising both RNA and PNA (see Supporting Information, SI† ). In order to obtain information on strand stoichiometry in this hybrid complex, we subjected it to Nano-Electrospray Ionisation mass spectrometry (NanoESI-MS, Micromass Q-TOF Ultima). An equimolar solution of P and R at 0.4 mM each in 100 mM NH4OAc, pH 4.5 were annealed and analyzed by positive ion nano-ESI-MS.6 Upon injection of the complex, the initial spectrum showed clearly a broad peak centered at m/z 1297.97 (Fig. 2A) where, the associated peak separation of (0.16 ± 0.03 m/z units) indicated that this was due to a hexa charged species. This corresponds to an associated molecular weight of 7787.82 ± 1 Da which is consistent with a four stranded entity [2MR + 2MP + 10H+ + 2NH4+]6+ (computed molecular weight. 7786.28 ± 1 Da) that is composed of two strands each of P and R with 10 additional protons. Interestingly, due to the high source temperature employed, the tetramer started dissociating with time and the broad peaks in Fig. 2B gradually disappeared at the expense of a family of new peaks also centered at the same m/z regime (see Fig. 1B). These peaks are equidistant with a constant separation of 7.6 ± 0.3 m/z units indicating that they correspond to multiply sodiated forms of triply-charged species of m/z 1299.35, with an associated molecular weight 3898.05 ± 1 Da. This in good correspondence with a dimeric entity composed of R and P, namely, [MR + MP + 5H+ + Na+]3+ (computed molecular weight 3899.15 ± 1 Da). This fragmentation pattern supports a model where the tetrameric complex is composed of two identical RP heterodimeric subunits each of which is held together by 5 additional protons. Furthermore, the tetramer evidenced a cooperative thermal transition characteristic of hemiprotonated C–C+base pairs, as found in i-motifs, by UV spectrophotometry at 295 nm (Fig. 3). Thus at pH 4.5, R and P in a 1 : 1 ratio forms a tetramer composed of two identical RP heterodimers held together by C–C+base pairs.
 | ||
| Fig. 1 Structure of the labels used in the study. Chart shows the sequences of RNA and PNA used. | ||
![Partial nano-ESI MS spectrum of an equimolar mixture of R and P, showing peaks corresponding to (A) a hexacharged tetramer [2MR + 2MP + 10H+ + 2NH4+]6+ and (B) Triply charged, multiply sodiated, dimer [MR + MP + 5H+ + Na+]3+ where MR and MP are the molecular masses of R and P respectively.](/image/article/2008/CC/b713525d/b713525d-f2.gif) | ||
| Fig. 2 Partial nano-ESI MS spectrum of an equimolar mixture of R and P, showing peaks corresponding to (A) a hexacharged tetramer [2MR + 2MP + 10H+ + 2NH4+]6+ and (B) Triply charged, multiply sodiated, dimer [MR + MP + 5H+ + Na+]3+ where MR and MP are the molecular masses of R and P respectively. | ||
 | ||
| Fig. 3 UV denaturation profile at 295 nm of P4, R2P2 and D2P2 complexes at comparable strand concentration in 30 mM Acetate buffer, pH 4.5. | ||
To elucidate the strand polarity in the R2P2 hybrids, self-quenching experiments (SpexFluorolog-1) were performed by forming hybrids with fluorescently labelled tetramethylrhodamine (TMR) derivatives of R and P. TMR self quenches due to exciton coupling with an R0 of 44 Å.7 Thus in an R2P2 complex, where one of the components is TMR-labeled, fluorescence quenching should provide an insight as to the relative like strand polarities. The observed distances obtained from all the possible combinations of labelled R and P are tabulated in Table 1.
| Labelled Hybrid complexes | Calc. Distancea /Å | Calc. IntensitybFDAcalc) | Obs. Intensity (FDAobs) | Obs. Distance/Å | r |
|---|---|---|---|---|---|
| a Distances were calculated from models of the hybrid RNA2-PNA2 i-motifs constructed using PyMOL software based on the NMR structure parameters of the RNA4 i-motif.10 Interfluorophore distances Rcalc, were measured incorporating the linker connecting the fluorophores. The model used the linkers in an all-trans conformation, taking the furthest interfluorophore distance. b Expected intensity (FDAcalc), based on the calculated quenching efficiency (E), for the calculated distance Rcalc, from the experimentally obtained FD. Distance accuracies are limited by the linker lengths of the freely rotating fluorophores represented in the error bar. All experiments were performed in triplicate and the average values are presented. | |||||
| N-TMR-P + R | ∼48 | 2.6 × 104 | 1.8 × 104 | 43 ± 5 | 0.04 |
| 3′-TMR-R + P | ∼45 | 6.4 × 104 | 8.4 × 104 | 50 ± 5 | 0.04 |
| 3′-TMR-R + N-Dabcyl-P | ∼19 | 4.6 × 103 | 4.2 × 103 | 19 ± 5 | 0.07 |
| 3′-TMR-R + C-Dabcyl-P | ∼23 | 2.3 × 105 | 3 × 105 | 24 ± 5 | 0.07 |
In the hybrid incorporating 1 : 1 P : 3′-TMR-R (20 µM), the interfluorophore distance was found to be 50 Å which corresponded to an antiparallel alignment of RNA strands in the tetramer. Similarly, in the hybrid comprising 1 : 1 N-TMR-P : R (10 µM), an interfluorophore distance of ∼43 ± 5 Å was obtained, revealing that the PNA strands are also arranged antiparallel to each other. Both R–P heteroduplexes can intercalate in two different configurations. In model 1 (Fig. 3) one narrow groove has both the RNA strands and the other, both the PNA strands. In Model 2, both narrow grooves have one RNA and one PNA backbone each. To address this, we formed a hybrid i-motif comprising 1 : 1 3′-TMR-R : N-Dabcyl-P (20 µM). Theoretical estimates of these interfluorophore distances, taking into account linker lengths and assuming groove dimensions commensurate with RNA4 i-motifs, were found to be ∼19 Å and ∼9 Å for models 1 and 2 respectively. A quenching efficiency of ∼85% was obtained indicating a TMR-Dabcyl separation of ∼19 ± 5 Å, consistent with Model 1 (Fig. 4A). Quenching measurements on 1 : 1 3′-TMR-R and C-Dabcyl-P (20 µM) yielded a distance of ∼24 Å (Rcalc was ∼23 Å) also consistent with Model 1.
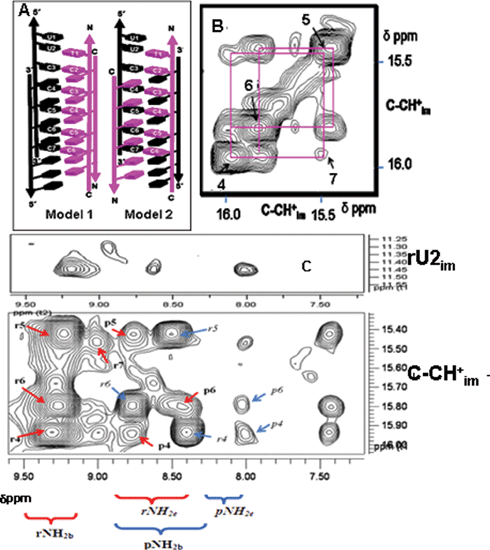 | ||
| Fig. 4 (A) Two possible models of the RP hybrid i-motif. Model 1: Both RNA strands (purple) are in one narrow groove. Model 2: One RNA and one PNA strand (black) in each narrow groove. Two dimensional NOE spectrum of the R2P2 hybrid i-motif at 1 mM strand concentration at 4 °C (200 ms mixing time). (B) NOE connectivities between four cytidine imino protons are shown, RNA residues (1–7) are indicated by the corresponding number. (C) Correlation of the NH2b, NH2e, with the imino proton (NHim) region and rU2im. The crosspeaks of the RNA NH2b protons of the rC–pC base pairs are labelled in bold r4, r5, r6, r7etc., while the corresponding RNA NH2e protons are labelled in italics r4, r5, r6. Similarly crosspeaks of PNA NH2b protons of rC–pC are labelled in bold p4, p5, p6, while the corresponding PNA NH2e protons are labelled in italics p5, p6. | ||
To confirm the findings from fluorescence quenching measurements and to obtain proof of RP heteroduplex intercalation characteristic of i-motifs, 1 mM R2P2, 30 mM d3-NaOAc, pH 4.5 was investigated by 1D and 2D NMR spectroscopy (Bruker AV 700 MHz). The 1D spectrum showed peaks in the region 15–16 δppm (SI), characteristic of the imino protons found in C–H+C base pairs, confirming the findings from NanoESI-MS and UV melting data.8 NOESY experiments on the R2P2 complex showed several crosspeaks. Two distinct regions characteristic of RP heteroduplexation and intercalation respectively are shown. These are (i) the imino proton (NHim)–amino proton (NH2b and NH2e) regions (Fig. 4C) and (ii) the NHim–NHim region. Cytosine imino protons (NHim) showed two sets of crosspeaks each to the hydrogen bonded (NH2b) and non-hydrogen bonded amino protons (NH2e). One set corresponded to the NH2b and NH2e protons of the RNA residues which were identified from their connectivities with rU2 while the other set corresponded to the NH2b and NH2e of the PNA residues, identified by their connectivity with pT1(CH3) and backbone methylenes (data not shown). Given that only a single set of U2im–C3NH2b, U2im–C3NH2e and U2im–CH6 crosspeaks were observed (Fig. 4C, lower panel) this confirmed the existence of only a single conformer of R2P2 i-motif in solution. This proves that the CH+C base pairs are asymmetric. The formation of rCH+–pC base pairs confirms heteroduplexation, which is in excellent correlation with the NanoESI-MS and fluorescence quenching experiments.
Intercalation of the heteroduplexes was proved by NOE crosspeaks shown by the rCn–(NHim)–pC(n–1) protons (Fig. 4B).9 The stacking order was identified as rU1–rU2–rC3–rC7–rC4–rC6–rC5–rC5–rC6–rC4–rC7–rC3 in one minor groove and pC2–pC6–pC3–pC5–pC4–pC4–pC5–pC3–pC6–pC2 in the other minor groove. Furthermore the observation of ribose sugar –sugar contacts evidenced by the H1′–H1′ as well as H1′–H2′ crosspeaks (ESI† ) indicates that both the ribose containing strands are in very close proximity, at a distance similar to that found in RNA4 i-motifs, confirming that both RNA strands flank a single narrow groove as in Model 1. All observed NOE connectivities are summarized in the schematic (ESI† ). The structure of the R2P2 i-motif is analogous to the ‘M’ form of RNA4 i-motifs.10 In the R2P2 hybrid, one potential intercalation site between rU2 and rC3 is empty. The intercalation topology of the hybrid reflects the optimization between maximizing the stabilizing sugar -sugar contacts and minimizing the destabilizing 2′-OH steric clash in a background of constant electrostatics in a given narrow groove environment. Thus, the partially intercalated topology, reveals that i-motif intercalation topology is highly sensitive to narrow groove interactions, where, the elimination of a single destabilizing 2′-OH interaction completely favours this topology.10
We have created a new RNA -based non-Watson–Crick building block with potential application in structural RNA nanotechnology. Via hybridization to DNA and RNA , PNA bearing functional moeities have been used for several applications such as genotyping, protein identification and biosensing.11 Thus, via the PNA component, this new buliding block has the scope to introduce a variety of moieties on a given RNA architecture to convert an inert RNA scaffold into a functional scaffold.12
Notes and references
- S. Pitchiaya and Y. Krishnan, Chem. Soc. Rev., 2006, 35, 1111 RSC; B. Samori and G. Zuccheri, Angew. Chem., Int. Ed., 2005, 44, 1166 CrossRef CAS; N. C. Seeman, Biochemistry, 2003, 42, 7259 CrossRef CAS; J. Bath and A. J. Turberfield, Nat. Nanotechnol., 2007, 2, 275 Search PubMed.
- Y. Krishnan-Ghosh, D. Liu and S. Balasubramanian, J. Am. Chem. Soc., 2004, 126, 11009 CrossRef CAS; Y. Krishnan-Ghosh, E. Stephens and S. Balasubramanian, Chem. Commun., 2005, 5278 RSC; S. Modi, A. H. Wani and Y. Krishnan, Nucleic Acids Res., 2006, 34, 4354 CrossRef CAS; H. B. Ghodke, R. Krishnan, K. Vignesh, G. V. P. Kumar, C. Narayana and Y. Krishnan, Angew. Chem., Int. Ed., 2007, 46, 2646 CrossRef CAS.
- L. Jaeger and N. B. Leontis, Angew. Chem., Int. Ed., 2000, 39, 2521 CrossRef CAS; B. Liu, S. Baudrey, L. Jaeger and G. C. Bazan, J. Am. Chem. Soc., 2004, 126, 4076 CrossRef CAS; A. Chworos, I. Severcan, A. Y. Koyfman, P. Weinkam, E. Oroudjev, H. G. Hansma and L. Jaeger, Science, 2004, 306, 2068 CrossRef CAS.
- R. T. Batey, R. P. Rambo and J. A. Doudna, Angew. Chem., Int. Ed., 1999, 38, 2326 CrossRef; N. Ban, P. Nissen, J. Hansen, P. B. Moore and T. A. Steitz, Science, 2000, 289, 905 CrossRef CAS.
- K. Gehring, J. L. Leroy and M. Guéron, Nature, 1993, 363, 561 CrossRef CAS; K. Snoussi, S. Nonin-Lecomte and J. L. Leroy, J. Mol. Biol., 2001, 301, 139 CrossRef; A. J. Brazier, J. Fisher and R. Cosstick, Angew. Chem., Int. Ed., 2006, 45, 114 CrossRef; U. Diederichsen, Angew. Chem., Int. Ed., 1998, 37, 2273 CrossRef CAS.
- Single stranded P shows a molecular mass (MP) of 1795.8 Da, while single stranded R shows an associated mass (MR) of 2075.3 Da. These values were taken to compute molecular weight of the complex.
- S. Bernacchi and Y. Mély, Nucleic Acids Res., 2001, 29, e62 CrossRef CAS.
- J. L. Mergny, L. Lacroix, X. Han, J. L. Leroy and C. Helene, J. Am. Chem. Soc., 1995, 117, 8887 CrossRef CAS.
- In this region, however, we were unable to detect the rC3+–pC2 base pair which may be broadened out due to fast exchange as seen in the case of short-lived external CH+–C base pairs.
- RNA4 i-motifs show two populations, partially and fully intercalated topologies in a 75 : 25 ratio. ( K. Snoussi, S. Nonin-Lecomte and J. L. Leroy, J. Mol. Biol., 2001, 301, 139 Search PubMed ).
- B. S. Gaylord, M. R. Massie, S. C. Feinstein and G. C. Bazan, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 34 CrossRef CAS; J. Zielinski, K. Kilk, T. Peritz, T. Kannanayakal, K. Y. Miyashiro, E. Eiríksdóttir, J. Jochems, Û. Langel and J. Eberwine, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 1557 CrossRef CAS; B. Liu and G. C. Bazan, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 589 CrossRef CAS.
- We thank the NanoScience and Technology Initiative of the Department of Science and Teschnology, Govt. of India and National Facility for High Field NMR, TIFR. SC and SM thank CSIR, Govt. of India for Junior Research Fellowships. YK thanks the Department of Biotechnology for the Innovative Young Biotechnologist Award.
Footnote |
| † Electronic supplementary information (ESI) available: Native PAGE, Circular Dichroism, UV melts, Fluorescence titration and raw data, 1D and 2D NMR. See DOI: 10.1039/b713525d |
| This journal is © The Royal Society of Chemistry 2008 |
