Hot off the press
Hot off the Press highlights recently published work for the benefit of our readers. Our contributors this month have focused on the imaging of cell surface glycans, ubiquitin’s dual role in the regulation of SRC-3 functional life time and the X-ray crystallisation of recombinant rhodopsin. New contributors are always welcome. If you are interested please contact molbiosyst@rsc.org for more information, we’d like to hear from you.
Imaging of cell surface glycans
Reviewed by: Ljiljana Fruk, University of Dortmund, Germany
One of the challenges in enabling the visualisation of biomolecules in living systems is finding the right approach to introduce the labels in such way so that they do not interfere with complex biological processes. In the meanwhile there are many strategies for fluorescent labelling of biomolecules, particularly proteins, available but many are not applicable to other classes like glycans or lipids. Incorporation of small functional groups in the target molecule using cells’ biosynthetic machinery proved to be an efficient way of circumventing some problems that arise when conventional fluorescent labelling is used. Carolyn Bertozzi and colleagues used this approach, the bioorthogonal chemical reporter technique, to image glycan on the cell surface with multiple labels. Azides and ketones were used as small functional group tags to which fluorescent labels could be covalently attached (Fig. 1). First, azides were introduced into cell surface glycan by metabolic labelling using the N-azidocetylmannosamine precursor for sialic acid. The cells bearing azido sialic acid were then treated with different phosphine modified fluorescent dyes to undergo Staudinger ligation and Cy5.5 phosphine showed superior fluorescence and low background labelling. Additionally, the multiple labelling was attempted which could enable simultaneous monitoring of different glycan subtypes. Cells were grown in presence of different metabolites to introduce azide and ketone groups to two different sugars on the same cells. Ketones react readily with aminoxy or hydrazide moieties and therefore dual labelling could be achieved by using appropriate dye derivatives. This was indeed the case and different types of glycans, sialic acid and GalNAc residues, have been visualised simultaneously on the same cell which could, in the future, ease the monitoring of glycan expression and dynamics.
P.V.Chang, J. A. Prescher, M. J. Hangauer, C. R. Bertozzi, J. Am. Chem. Soc., 2007, 129, 8400–8401.
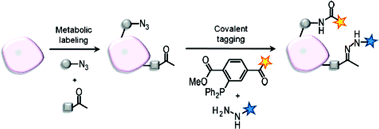 | ||
| Fig. 1 Strategy for dual imaging of azide- and ketone-labeled cell surface biomolecules. Figure used with permission from J. Am. Chem. Soc., 2007, 129, 8400–8401. Copyright 2007, American Chemical Society. | ||
A dual role for ubiquitin in the regulation of SRC-3 functional life time
Reviewed by: Sahron Aviram, Division of Translational Research, Dept. of Internal Medicine, UT-Sowthwestern Medical Center.
A recent study from Bert O'Malley's lab (Cell, 129, 1125–1140) examined the turnover of the steroid receptor coactivator-3 (SRC-3), a bona fide oncogene, that interacts with and activates nuclear receptors and transcription factors. The data suggest a dual role for ubiquitin in both activating SRC-3, and mediating its proteasomal degradation. These roles for ubiquitin were previously suggested independently to be important for regulating the activity of transcription factors.
In trying to understand how cellular signaling regulates SRC-3 turnover, and how it affects SRC-3 transcriptional activity, the authors characterized the degradation pathway of SRC-3. Their data indicate that phosphorylation of SRC-3 stimulated by estradiol, is a prerequisite step for SRC-3 ubiquitination by the SCFFbw7α and proteasomal degradation. Their most interesting finding is the identification of a relatively stable and low molecular weight ubiquitin modified SRC-3, which they show to be multi-(mono) ubiquitinated. This derivative, as well as the polyubiquitinated activator, were strongly detected in the presence of Fbw7α, suggesting that both multi-(mono) ubiquitination and polyubiquitination of SRC-3 are catalyzed by the same ubiquitin ligase.
Upon further assessment of the role that SRC-3 turnover plays in transcription activation, the authors demonstrate that SRC-3 phosphorylation and Fbw7α dependent ubiquitination both enhanced the interaction with nuclear receptor and DNA. These suggest that ubiquitintion of SRC-3 can also act as an “actron” and activate SRC-3 functions.
Comparison of the ability of multi-(mono) ubiquitinated versus polyubiquitinated SRC-3 to activate transcription clearly showed that mono-ubiquitination of SRC-3 is sufficient for full activation of transcription, and can recruit RNA PolII. Moreover, SRC-3 polyuibiquitination was shown to be inhibited by interrupting transcription with actinomycin D. These data strongly suggested that ubiquitination of SRC-3 is a biphasic event, where mono-ubiquitination and polyubiquitination take place at different stages during transcription. Considering the unique role that these modifications play, the author suggested a model in which SRC-3 functional life time is regulated by a ubiquitin clock.
Ubiqutinaion of transcription factor and activators is known to be important for the regulation of transcription. However the majority of the transcription activators were shown to be either regulated by non-proteolytc ubiquitination (mono-ubiquitination) or proteolytic ubiquitination.
Since these modifications appear to act independently of one another, they are thought to be catalyzed by different ubiquitin ligases. This work expends our understanding of the role that ubiquitin play in activation of transcription, and probably in other signaling pathways.
Ray-Chang Wu, Qin Feng, David M. Lonard, and Bert O'Mally, SRC-3 Coactivator Functunal Lifetme Is Regulated by a Phospho-Dependent Ubiquitin Time Clock, Cell, 2007, 129, 1125–1140.
Rhodopsin shows the way
Reviewed by Kutti R. Vinothkumar, MRC-LMB, Cambridge, UK.
Structural determination of membrane proteins, in particular the proteins of eukaryotic origin has often been difficult due to their instability in detergent micelles and low natural abundance. A recent flurry of genomics has allowed identification of prokaryotic homologues of many important classes of membrane proteins such as transporters, channels and many others, leading to a structure based understanding of many fascinating proteins.
However, the main family of eukaryotic seven trans-membrane domain membrane proteins, the so-called G-protein coupled receptors (GPCRs), have eluded many decades of crystallization and unfortunately, no prokaryotic homologues have been found yet. The human genome contains several thousand different GPCRs that play major roles in biologically important signalling pathways and many of them are prime candidates for drug targets in the treatment of various diseases.
To date, the only known structure of a GPCR is that of rhodopsin. The high abundance of rhodopsin in the retina and its higher stability in the presence of a covalently bound chromophore in its ground state are the primary reasons for obtaining good reproducible crystals suitable for high-resolution crystallography. In contrast, very few GPCRs have been expressed at the high levels suitable for crystallography and their stability proves to be a major obstacle in obtaining crystals.
Now, the group of Gebhard Schertler and Daniel Oprian have expressed rhodopsin recombinantly in mammalian cells and obtained a structure, which is very similar to native rhodopsin. How did they achieve this? Using the vast biochemical data available for rhodopsin and a clever structure based design they were able to stabilize the recombinant protein by introducing a disulphide bond, which gives a remarkable increase in thermal stability with or without the ligand bound. Importantly, this modification did not compromise the ability of rhodopsin to bind its ligand or activate the G-protein.
Another remarkable feat achieved is obtaining the structure from a crystalline needle that measures just 5 × 5 × 90 μm3. Using a 5 μm X-ray beam and micro-crystallography, images were collected at different positions of the crystal by translation of the needle, allowing maximum data to be collected with minimal radiation damage and background. The structure is essentially similar to native rhodopsin with little functional information gained but the ability to obtain sufficient amounts of membrane protein from mammalian cells for X-ray crystallography, the first of its kind, opens up a new path in understanding mutants that affect the function of rhodopsin and the signalling cycle. Similar strategies to introduce stabilizing mutants of other GPCRs is possibly the way to go and will help tackle one of the final frontiers in the field of membrane proteins.
Standfuss J., Xie G., Edwards P.C., Burghammer B., Oprian D.D., Schertler F.X.G., J. Mol. Biol., DOI: 10.1016/j.jmb.2007.03.007
Hot off the RSC Press
Molecular gate with a silver key
Reviewed by: Clare Boothby, Royal Society of Chemistry, Cambridge.
French scientists reveal the design of a new molecular gate locked by a silver ion.
The gate was designed by a team at Louis Pasteur University, Strasbourg. As well as providing a controllable component for use in molecular machines, the researchers see a future for it as a switch that can store and release ions.
The gate (see Fig. 2) has a porphyrin hinge and a freely rotating handle. The hinge and handle both include a pyridyl group designed to form a complex with a silver ion. When no silver ions are around, the handle can rotate freely around the hinge. As soon as a silver ion enters the gate, however, it forms a complex with both pyridyl groups, locking the handle in place.
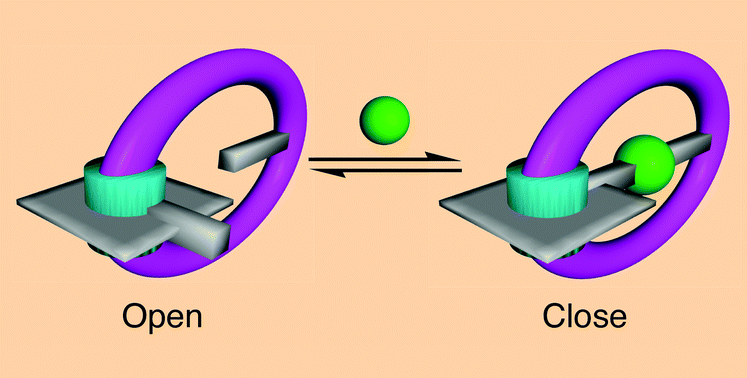 | ||
| Fig. 2 The metal key (green) locks the gate with coordination bonds. Copyright 2007, Royal Society of Chemistry. | ||
The inspiration for the system came from several places, including public park gates and ATP synthase, an enzyme which acts as a rotating molecular motor, said Wais Hosseini, who led the research.
‘The design principle is interesting because it is rather general and opens the way to set-up many other systems. In particular, we are currently working on other molecular gates based on different types of coordination sites and other metal centres such as mercury or palladium. We're also working on a system operated by proton concentration,' said Hosseini.
‘The final goal of our investigation is to open the path to rotational motors with control of the direction of rotation,' he added. 'We are currently working on how to introduce a potential gradient to achieve that.'
Aurélie Guenet, Ernest Graf, Nathalie Kyritsakas, Lionel Allouche and Mir Wais Hosseini, Chem. Commun., 2007, 2935
Crystals as genes?
Reviewed by: Caroline Moore, Royal Society of Chemistry, Cambridge.
The hypothesis that crystals could have been primitive genetic materials has been put to the test by US scientists.
Nearly three decades ago, Graham Cairns-Smith proposed that the first genetic systems must have been more primitive than the sophisticated chemistries of DNA and RNA. He argued that crystals, particularly clay minerals, have the capacity to act as primitive genes.
His idea was that imperfect crystals can act as genes by transferring information from one crystal to another by means of their imperfections. Screw dislocations, for instance, are often replicated through crystal growth, so the arrangement of these dislocations in a crystal can be considered a store of information. This so-called ‘crystals-as-genes' hypothesis has captivated the imagination of scientists for many years, said Bart Kahr from University of Washington, Seattle, US, but it has never been put to the test, until now.
Kahr and colleagues have designed the first experiment to examine the idea that crystals can act as a source of transferable information, using crystals of potassium hydrogen phthalate.
‘Our paper is really an experimental test of an idea whether or not that idea is relevant to life's origins,' said Kahr.
The researchers looked at the distribution of hillocks in the crystal system. ‘Mother' crystals with these imperfections were cleaved with a razor and used as seeds to grow ‘daughter' crystals from solution. The resulting daughter crystals were imaged using fluorescence microscopy to see whether the spatial distribution of the hillocks had been inherited.
Kahr found that, as Cairns-Smith suggested, the distribution of the crystal defects can be transferred from one crystal to another. However, a large number of new hillocks, ‘mutations', were also observed (Fig 3). For crystals to resemble genes there must be more inheritance than mutation in successive generations.
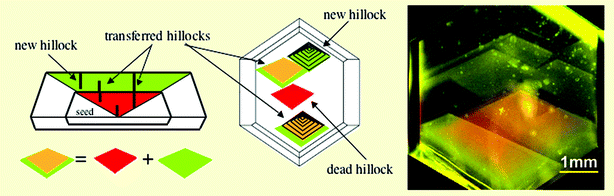 | ||
| Fig. 3 Crystal defects can be transferred but ‘mutations' were also observed. | ||
‘While we determined that the dislocations in the crystal system that we studied were not faithful enough to store and transfer information form one generation to the next,' said Kahr, ‘we did demonstrate how we can use luminescent molecules to identify the sequence in time and space of all growth active dislocations.'
Cairns-Smith himself is not deterred. ‘I would say that the success of [the] idea that RNA preceded DNA has provided inadvertent support for crystal genes,' he said. ‘The big thing missing... is an account of how activated nucleotides might have appeared on the primitive Earth as feedstock for replicating RNA molecules. The kind of organic chemical competence required here could only have been the result of natural selection—based of course on some other genetic material.'
Kahr, however, is more guarded. ‘We wanted to try to bring one aspect of the multifaceted proposal of Cairns-Smith to the realm of repeatable experimental science.'
‘I would hope that our experiment would encourage scientists to subject other aspects of the broad crystal-as-genes hypothesis to the scrutiny of experiment,' added Kahr.
While they may not have unravelled the mysteries of life's origins, the scientists have provided new tools for studying crystal growth mechanisms, an area of great interest in many aspects of materials science.
Theresa Bullard, John Freudenthal, Serine Avagyan and Bart Kahr, Faraday Discuss., 2007 DOI: 10.1039/b616612c
Clearing a path to cancer detection
Reviewed by: Wendy Crocker, Royal Society of Chemistry, Cambridge.
Improved imaging of prostate cancer proteins in single cells is possible thanks to scientists at the University of Manchester. The UK researchers identified and investigated an optical illusion in the imaging of single cells that gave inconsistent results.
‘Developing the use of infrared spectroscopy in cancer diagnosis' is the aim of the UK team led by Peter Gardner. ‘Prostate cancer, if it remains localised, is a manageable condition and patients can survive for many years,' Gardner said. ‘However, the cancer can spread, particularly to the bone, and this is almost always fatal.' Gardner's team, in collaboration with Noel Clark, a consultant urologist at the Christie Hospital and Paterson Institute for Cancer Research, have therefore been studying how cancer cells move through tissue.
They study cancer cells by sending a beam of infrared (IR) radiation through the cancer cell and what it is growing on (its biological support generally called the substrate). The beam then bounces off an IR reflective surface and reflects back into the detector, measuring how much is absorbed. A map of the absorption creates an image of the cancer cell proteins (Fig 4).
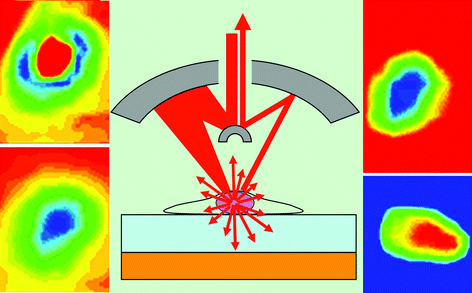 | ||
| Fig. 4 Some protein substrates give negative images of cancer cells. | ||
The problem seen by Gardner's team was that, with some high protein substrates, they sometimes saw negative images of the cells. This illusion was caused by the IR radiation being scattered by the nucleus of the cancer cell. As it scatters in all directions, some goes directly into the detector resulting in more radiation being recorded, indicating that less has been absorbed. This gives the illusion of a negative image of the cancer cell.
Deducing the origin of these optical illusions ‘contributes significantly to the debate concerning "anomalies" observed in infrared-microscopy mapping of single cells,' according to John Chalmers of VS Consulting, Stokesley, UK. The Manchester researchers cautioned that ‘people using infrared spectroscopy to study cells on biological supports must be careful how they interpret their results'.
Joe Lee, Ehsan Gazi, John Dwyer, Michael D. Brown, Noel W. Clarke, James M. Nicholson and Peter Gardner, Analyst, 2007, 132, 750
| This journal is © The Royal Society of Chemistry 2007 |
