Hot off the Press
In the Hot off the Press section of Molecular BioSystems members of the Editorial Board and their research groups highlight recent literature for the benefit of the community. This month the highlighted topics include MALDI-TOF analysis of siRNA degredation published in this issue of Molecular BioSystems, one of our most regular contributers to Hot off the Press Ljiljana Fruk and an item published recently in one of the RSC’s journals.
Weighing in with the answers: insights into the mechanism of siRNA degradation by MALDI
This latest paper out of the group of Mike Gait is a very nice demonstration of the power of MALDI-TOF mass spectrometry to analyse the method of action of siRNA degradation. siRNAs are short, double stranded ribonucleic acids that are typically 19–20 bases in length. They have been used extensively to reduce gene expression of target RNA and can be prepared synthetically. This makes them very attractive as agents for regulating gene expression from a therapeutic perspective and also in terms of their ability to regulate gene expression to try and understand more complex biological interactions. However, there is, as ever, an issue with using a synthetic molecule in a living environment, namely in this case the degradation of the siRNA which reduces its activity. Approaches have been taken to try and circumvent this problem including modification of the ribonucleotides with modifications such as 2′-O-methyl. Until this study the approach to modification was less systematic and used more trial and error in achieving the required stability. Initially the authors identified that there is a problem when trying to identify the mechanism of cleavage of siRNA due to the lack of reliable and sensitive methods for this type of experimentation. MALDI-TOF offers significant advantages over other techniques such as gel electrophoresis and this paper demonstrates the power of the technique when applied to siRNA degradation studies.The group at the MRC are well known for studies into the delivery of therapeutic nucleic acid analogues into cellular environments. As such, part of this study focussed on the study of the breakdown of cell penetrating peptides and delivery reagents, such as cholesterol, conjugated to the siRNA. This data was also compared to data acquired by polyacrylamide gel electrophoresis to demonstrate that the breakdown was occurring in the siRNA and not the peptide or cholesterol. The mass spectral data indicated cleavage at a particular point in the sequence and convincingly demonstrated that the method of activity of the enzyme was based on a RNAse A type cleavage mechanism, i.e. degradation occurred internally within the siRNA as opposed to degradation from one of the terminals as in an exonuclease activity. The data obtained from the MALDI-TOF indicates that the cleavage occurs after pyrimidine nucleotide residues. The authors also say that the RNAse A like activity is unlikely to recognise and cleave double stranded RNA, and it is therefore probable that the cleavage only takes place when the siRNA duplex is breathing and a region of single stranded RNA is produced. This again fits in with the observation that the cleavage takes place at a pyrimidine base site and in particular the UpA region especially if at one end of the duplex. If the UpA sequence is near the centre it was found that the cleavage rate was much slower and did not appear to be any more vulnerable than other pyrimidines in the duplex. This is important as most siRNA duplexes have been designed to be either A or U rich at one end and this now allows appropriate modification of the pyrmidine to improve stability. This is a highly significant result as it is the first time that this has been demonstrated experimentally and it also indicates how siRNA can be made less susceptible to cleavage by careful positioning of modified ribonucleotides. If for instance a 2′-O-methyl group is used then at the cleavage site then the siRNA is rendered stable to degradation in the serum samples. The information gained from this study allows the careful positioning of the modified base at the enzyme cleavage site as opposed to a blanket coverage of modification of pyrimidine bases and will have significant impact on the ability of siRNA to exist in complex biological systems. This is an important advance in the understanding of siRNA stability and the mechanism of degradation in serum and will have significant impact on the future use of siRNA as a potential therapeutic agent.
John J. Turner, Simon W. Jones, Sterghios A. Moschos, Mark A. Lindsay and Michael J. Gait Molecular BioSystems, 2007, 3(1), 43–50.
Reviewed by: Duncan Graham, University of Strathclyde.Profile of Hot off the Press Contributor Ljiljana Fruk
Ljiljana graduated from University of Zagreb, Croatia and completed her PhD in biospectroscopy at the University of Strathclyde, Glasgow, in the group of Prof. Ewen W. Smith. She came to Christof Niemeyer's lab at the University of Dortmund, Germany as a Humboldt Fellow and is now in the final stage of a two year Marie Curie Postdoctoral Fellowship. Her work focuses on the development of a novel class of addressable redox biocatalysts: well-defined semisynthetic DNA–heme enzyme conjugates. She is exploring ways of using DNA strands to immobilize or spatially organize enzymes to enable their catalytic activity and interaction with inhibitors or other molecules of interest to be investigated. One of the goals of this research is the development of an analytical platform that allows the high-throughput screening of novel catalytic properties of enzymes and the design of a range of different biosensors. She is very much interested in carrying out fundamental research on protein interactions as well as focusing on biosensor development and biospectroscopy.
More details on: http://www.chemie.uni-dortmund.de/groups/niemeyer/ljiljana/indexljiljana.html
Novel activity of engineered aldolase
Enzyme engineering has led to the design of ranges of different enzymes with novel or improved activities and many of them have been used as a tool in large scale chemical synthesis. Aldolases are particularly interesting group of enzymes because of their ability to make (and break) enantioselective C–C bonds. The group of Donald Hilvert from ETH Zürich has previously shown that single active site mutation (Y265A) in pyridoxal 5′-phosphate (PLP) dependent alanine racemase (alrY265A) can increase retro-aldolase activity against phenylserine (Scheme 1) by five orders of magnitude.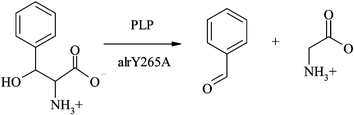 | ||
| Scheme 1 Reprinted with permission from F. P. Seebeck, A. Guainazzi, C. Amoreira, K. K. Baldrige, D. Hilvert, Angew. Chem., Int. Ed. 2006, 45, 6824–6826. | ||
Now the same group has shown that this engineered enzyme catalyses also retro-aldol reaction of α-substituted β-phenylserines with high stereoactivity. This type of reaction involving quaternary α-amino acids is not observed in natural aldolase, but it has a huge potential in chemical synthesis of amino acids. To start with, two β-hydroxyl amino acids with additional α-substituents were synthesized (2a, 2b). In the presence of alrY265A this amino acids initially react with enzyme bound PLP (3) which is then followed by Cα–Cβ bond cleavage of the adduct. In subsequent steps the product intermediate is protonated and hydrolyzed (by Lys39 or water) to give glycine (or alanine) and regenerated catalyst. The kinetic parameters obtained for this reaction showed that substrate is consumed with higher catalytic efficiency than in case of (2R, 3S)-β-phenylserine. The data also indicated that there is poor steroechemical control at β, but very high at α carbon. Additionally, computational (docking and ab initio) methods were used to investigate substrate binding to the active site of the enzyme giving insight into possible additional optimization of the mutant. This exciting contribution to the field of biocatalysis may soon prove to be a very important route to the enantioselective synthesis of novel amino acids.
F. P. Seebeck, A. Guainazzi, C. Amoreira, K. K. Baldrige, D. Hilvert, Angew. Chem., Int. Ed. 2006, 45, 6824–6826.
Reviewed by: Ljiljana Fruk, University of DortmundUpdate
In last month’s Hot off the Press, the article ‘Global gene expression signatures for global chemical probe and drug discovery’ cited an in press article. This has now been published and can be found in Nat. Chem. Biol., 2006, 2(12), 689–700.Hot off the RSC Press
Device monitors expression
A ‘living cell array’ to monitor cell responses to drugs could lead to a greater understanding of liver disease and its treatment.Martin Yarmush and a team at Massachusetts General Hospital, Boston, US, have developed a microfluidic device that allows them to study gene expression, the production of proteins from genes, in living cells.
Previously, scientists looking at gene expression would have to make destructive measurements, said Yarmush. The cell would be broken up for analysis, giving a snapshot of its response to a stimulus, he explained. Long term responses had to be assembled from separate cell populations.
Using the microfluidic system, the team was able to monitor gene expression continually, a so-called dynamic study. By altering the genes to express fluorescent proteins and exposing the cells to different conditions, the team could measure the effects on gene expression as a change in fluorescence.
The group developed its array to help study liver biology and disease. Yarmush explained that ‘the technology might help us advance from building static models of disease states to dynamic models of disease processes.’ The method also opens ‘additional possibilities’ for screening potential drugs, optimising the timing and doses of combination therapies, and studying the mechanisms underlying cell damage and recovery, he said.
As well as the benefits from being able to study the living cell, the microfluidic array allows fast, high-throughput analysis which is cheaper than existing options. Yarmush added, ‘the platform has already dramatically increased experimental throughput 100-fold, and there is tremendous potential for further scaling.’
 | ||
| Fig. 1 The array can monitor the effects of drugs and disease on living cells. Reproduced from Lab Chip, 2006, DOI: 10.1039/b612516f by permission of The Royal Society of Chemistry. | ||
KR King, S Wang, D Irimia, A Javaraman, M Toner and ML Yarmush, Lab Chip, 2006, DOI: 10.1039/b612516f.
Reviewed by: Laura Howes, Royal Society of Chemistry, Cambridge.| This journal is © The Royal Society of Chemistry 2007 |

