Hot off the Press
In the Hot off the Press section of Molecular BioSystems members of the Editorial Board and their research groups highlight recent literature for the benefit of the community. This month the highlighted topics include structural organization of the 19S proteasome lid, a hybrid molecular probe for analysis of biological samples, a three-hybrid trap for quantitative proteome fingerprinting, improving the robustness of enzymes by nanogel encapsulation and some items published recently in the RSC’s journals.
Structural organization of the 19S proteasome lid
The Robinson group at Cambridge have recently elucidated some of the molecular contacts within the lid of the 19S proteasome. The 19S proteasome is the “cap” component of the 26S proteasome that can be dissociated under high salt concentrations into a base portion and a lid. The lid consists of 9 proteins involved in protein deubiquitination, yet, a high-resolution structure of the complex has not been obtainable to date. The Robinson group has pioneered an interesting form of electrospray mass spectrometry that maintains the non-covalent contacts of a multi-protein complex throughout the ionization and mass analysis process. These ions in turn reveal mass information concerning the complex organization, and depending on the strength of the ionization process, sub-complexes. Previously, most complexes analyzed by this methodology had to be reconstituted from recombinantly expressed proteins, however, given the relative abundance of the proteasome within the cell, it is possible to purify the sub-complex at sufficient amounts to analyze with this technique. The Robinson laboratory not only identified the mass of the entire lid complex of 9 proteins but they were also able to identify sub-complexes of the lid. Namely, Rpn 5, 6, 9, and 8 form a sub-complex with 8 at the center of the complex, and Rpn 6, Rpn 9 and Rpn 12 exist on the periphery of the lid complex. The MS technology also revealed that Sem1, Rpn 7, and Rpn 3 exist in a complex. Furthermore, 3 and 5 were demonstrated to bind to each other by chemical cross-linking using an amine reactive cross-linking chemistry connecting the two complexes (Fig. 1). These data raise several interesting questions. In particular, it would be interesting to analyze whether any of these sub-complexes are functional apart from the proteasome, and what cellular processes regulate alternative sub-complex formation. Importantly, this work further strengthens this approach as a viable method to gather information detailing macro-molecular structure, a vexing problem faced by most molecular biologists.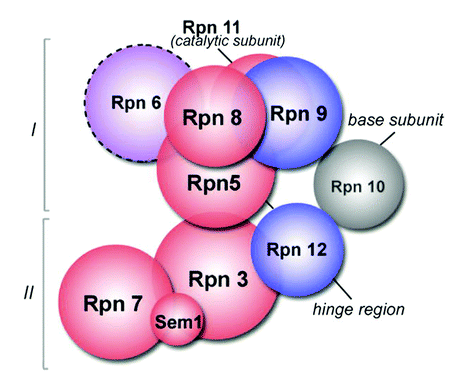 | ||
| Fig. 1 Structural organization of the 19S lid. Reproduced with permission from: Michal Sharon, Thomas Taverner, Xavier I. Ambroggio, Raymond J. Deshaies and Carol V. Robinson, PLoS Biol., 2006, 4(8), 1314–1323. | ||
M. Sharon, T. Taverner, X. I. Ambroggio, R. J. Deshaies and C. V. Robinson, PLoS Biol., 2006, 4(8), 1314–1323.
Reviewed by: Thomas Kodadek, University of Texas Southwestern Medical Center, Dallas, USA.Hybrid molecular probe for analysis of biological samples
Since the first paper describing their design and application was published in 1996, molecular beacons have attracted a huge interest in the scientific community and industry: becoming useful in a range of applications. The stem–loop design with dye pairs positioned on 5’ and 3’ ends have been improved over the course of recent years and used in live cell monitoring.Apart from DNA, molecular beacons were also used to monitor RNA in the cells. However, when used in vivo they often generate false positives due to degradation and protein binding. Now Weihong Tan’s group have designed a novel probe for monitoring RNA in vivo, which overcomes some of the limitations of the standard molecular beacons. They synthesised a probe called hybrid molecular probe (HMP) by linking two dye labelled single stranded DNAs (typically 12–25 bases) with a poly(ethyleneglycol) (PEG) polymer chain in such a way that different dyes (FAM and Cy5) were positioned on two different ends. Two ssDNA were complementary to adjacent areas of the target sequence and when hybridisation takes place 5’ and 3’ ends of the probe are brought in close proximity enabling FRET to occur (Fig. 2). This principle has often been used in numerous applications, but the novel feature of the probe is a PEG linker. As the authors mention they have chosen PEG because of the facile synthesis, variable length and good solubility in water. The linker also helps in keeping two DNA sequences together and enabling them to hybridise to one molecule instead of two. Additionally, PEG provides the scaffold for insertion of other functionalities to the probe e.g. biotin for immobilization or membrane penetrating peptide for probe delivery purposes. When compared to molecular beacons, HMP probe had a higher signal to noise ratio upon target binding, although molecular beacons showed slightly better single base mismatch discrimination. However, no protein interaction was observed in the case of HMP, while false positive signals appeared when molecular beacons were used. Finally, when both probes were tested in a cell lysate, HMP showed its superiority giving no false positives and responding to target sequences faster.
The robustness, selectivity and response time make this new type of molecular probe an excellent candidate for target detection in vivo and development of DNA/mRNA biochips.
 | ||
| Fig. 2 (Left) Schematic presentation of the working principle of a HMP. (Right) Hybridization of HMPTBL to its target sequence and its control in 20 mM Tris-HCl buffer (50 mM NaCl, 5 mM MgCl2, pH 7.5). Reproduced with permission from J. Am. Chem. Soc., 2006, 128, 9986–9987. Copyright 2006 American Chemical Society. | ||
C. J. Yang, K. Martinez, H. Lin, W. Tan, J. Am. Chem. Soc., 2006, 128, 9986–9987.
Reviewed by: Ljiljana Fruk, Universität Dortmund, Germany.MASPIT: Three-hybrid trap for quantitative proteome fingerprinting of small molecule–protein interactions in mammalian cells
Small molecules have diverse biological activity and are commonly employed as tools to study protein function in addition to their potential as drug candidates. Recent studies have illustrated the difficulty in predicting the global target profiles of small molecules demonstrating the importance of evaluating the mechanism of action of a small molecule on a proteome-wide scale. The authors have created a novel platform for studying protein–small molecule interactions within mammalian cells, which they term a mammalian small molecule protein interaction trap (MASPIT).The MASPIT assay relies on a chimeric cytokine receptor-associated JAK/STAT signaling system that employs an Escherichia coli DHFR (eDHFR) fusion protein. This is used for binding to hybrid small molecules that are covalently attached to methotrexate (MTX) via a polyethylene glycol repeat spacer. The eDHFR fusion protein binds tightly to the MTX portion of the small molecule hybrid. A number of small molecule hybrids were selected from diverse small molecule chemotypes. Proteins known to bind to these small molecules were developed with gp130 fusion proteins integral to the JAK/STAT signaling system. Therefore, when the protein of interest interacts with the small molecule ligand the gp130 fusion protein comes within proximity of the cytokine receptor chimera and stimulates the signaling event.
The MASPIT assay provided a method for cDNA library screening and target identification. MASPIT gives a high level of confidence with which small molecule–protein interactions can be characterized. Therefore, the authors were able to determine through a cDNA screen a kinase family inhibitor (EC50 of 0.028 µM for ABL kinase) with a core structure that could potentially be used in building an ephrin receptor tyrosine kinase inhibitor. These kinases have been implicated in the regulation of numerous biological processes.
The MASPIT system has the potential to provide proteomic selectivity profiles of small molecules based upon their mechanism of action. Furthermore, this system may be useful for determining novel therapeutic/clinical applications of small molecules.
M. Caligiuri, L. Molz, Q. Liu, F. Kaplan, J. P. Xu, J. Z. Majeti, R. Ramos-Kelsey, K. Murthi, S. Lievens, J. Tavernier and N. Kley, Chem. Biol., 2006, 13(7), 711–722.
Reviewed by: Aaron Wright, The Scripps Research Institute, California, USA.Improving the robustness of enzymes by nanogel encapsulation
Enzymes are playing a significantly increasing role in organic synthesis particularly in larger industrial processes. However, most of the time it is difficult to maintain their desired activity under sometimes harsh conditions of industrial biotechnology. This mainly refers to higher temperature and the presence of organic solvents which leads to the inactivation of sensitive biomolecules.A number of solutions have been tested to improve robustness of the enzymes, among them encapsulation in silica or polymer particles. Unfortunately this usually imposes steric constraints upon the molecule and hinders the substrate accessibility. In a recent paper, M. Yan and coworkers introduced a new method based upon the formation of single enzyme capsules with permeable coating. They took horseradish peroxidase (HRP)—an enzyme which is widely used in bioassays and biosynthesis—as a model. First they treated HRP with N-acryloxysuccinimide under mild alkali conditions to generate surface vinyl groups. Then in situ polymerisation was conducted using acrylamide as the monomer and a standard linker and initiator procedure. This has led to the formation of a polymer coated enzyme (Fig. 3). TEM studies proved that single enzymes are encapsulated giving spherically shaped nanogel particles. Interestingly, and most importantly, the activity of such enzyme nanogels did not decrease and only slightly differed from the kinetics of the native HRP. Additionally, thermal stability of the enzyme improved significantly. While free HRP started to experience the loss of activity at 40° C nanogel remained stable until 65° C and maintained 80% of activity after 90 min at this temperature. Additionally, tests with organic solvents showed that nanogel–HRP retains 80% of activity after solvent exposure while the native one is completely deactivated.
The described method of enzyme stabilisation is simple and effective. Although HRP is a very interesting enzyme, it still remains to be tested if this procedure is transferable to other enzymes. In any case, this novel HRP nanogel might soon prove to be important for a range of applications.
 | ||
| Fig. 3 Encapsulation of single protein in nanogel. Reproduced with permission from J. Am. Chem. Soc., 2006, 128, 11008–11009. Copyright 2006 American Chemical Society. | ||
M. Yan, J. Ge, Z. Liu and P. Quyang, J. Am. Chem. Soc., 2006, 128, 11008–11009.
Reviewed by: Ljiljana Fruk, Universität Dortmund, Germany.Hot off the RSC Press
Early detection of cancer cells
A way to detect cancer by monitoring proteins could lead to earlier diagnosis and faster treatment for patients, says a scientist in the US.Changes in the levels of proteins found in cells, known as the proteomic pattern, can be used to diagnose several diseases. However, monitoring this pattern is not an easy thing to do, said Gordon Whiteley at the National Cancer Institute at Frederick. Whiteley has highlighted the possibilities that advances in proteomic technologies now present. In particular, combining established techniques such as mass spectrometry, microarrays and computer informatics could lead to ‘innovative’ methods of diagnosis, said Whiteley (Fig. 4).
 | ||
| Fig. 4 Use of mass spectrometry, microarrays and computer informatics in diagnosis. Reproduced from Mol. BioSyst., 2006, 2, 358–363 by permission of The Royal Society of Chemistry. | ||
The early diagnosis of cancer is an important goal for researchers since timely treatment is often essential to combat the disease. Until recently, tests used to detect and monitor cancer have been limited to either imaging or immunoassay tests. Often these do not have the sensitivity to detect small changes in cells that can provide early evidence for cancerous growth, said Whiteley.
One of the key developments in the area, Whiteley added, has been the combination of two powerful techniques already used in the lab: laser capture microdissection and surface enhanced laser desorption ionisation. Using these tools, a pattern for ovarian cancer has been used to identify patients with cancer with 100 per cent sensitivity and 95 per cent specificity.
Other combinations of techniques are also proving useful in identifying patterns for several cancer related diseases, said Whiteley. ‘All of this needs enormous development but the results for patient care will be substantial.
G. R. Whiteley, Mol. BioSyst., 2006, 2, 358–363.
Reviewed by: Sarah Dixon, Royal Society of Chemistry, Cambridge, UK.Gold delivery kills cancer cells
Scientists in the UK have found a way to target cancer with gold.David Russell and co-workers at the University of East Anglia have designed a system that delivers a light sensitive drug to cancer cells. The system uses gold nanoparticles, to which the drug and another organic molecule, called a phase transfer agent, can be bound. This drug–nanoparticle complex can be used to kill cancer cells in a process called photodynamic therapy (PDT).
PDT is an established cancer treatment that is available in several countries. It uses the combination of a photoactive drug (a photosensitiser) and light. When stimulated by visible light the drug damages the target cancer cells, leaving healthy tissue unharmed.
Russell's system used a cancer drug based on a phthalocyanine molecule. These molecules are more specific to cancer tissue than others commonly used in PDT. However, this type of drug is not water soluble, making it more difficult to deliver it to target sites, said Russell. Using a phase transfer agent can help overcome this problem by making the drug–nanoparticle complex soluble in water, he explained
On irradiating with red light, the phthalocyanine drug produces an active form of oxygen, singlet oxygen, which is toxic to cells. The group found that the drug–nanoparticle complexes produced 50 per cent more singlet oxygen than the drug alone (Fig. 5)
 | ||
| Fig. 5 Combined confocal fluorescence and DIC images of HeLa cells following incubation with phthalocyanine–nanoparticle conjugates. The presence of the nanoparticle conjugates within the HeLa cells can be readily seen from the fluorescence emission (red) following excitation at 633 nm. Reproduced from Photochem. Photobiol. Sci., 2006, 5, 727–734 by permission of The Royal Society of Chemistry. | ||
Russell showed that the drug–nanoparticle complexes were taken up by cervical cancer cells in vitro. These cancer cells then died by apoptosis, or cell suicide. ‘While our results are very encouraging, the next key phase of the work is to take the in vitro study to the in vivo environment. To do this we are collaborating with a PDT group in Padova,’ said Russell.
David Phillips, a PDT expert from Imperial College London, UK, said, ‘This is a very useful, novel means of getting photosensitisers into cells, and in this case, the use of the gold nanoparticles enhances the singlet oxygen yield, so is a real bonus.’ Phillips suggests that, although there will be several factors to consider, the delivery system ‘promises well for future applications in vivo.’
M. E. Wieder, D. C. Hone, M. J. Cook, M. M. Handsley, J. Gavrilovic and D. A. Russell, Photochem. Photobiol. Sci., 2006, 5, 727–734.
Reviewed by: Katherine Vickers, Royal Society of Chemistry, Cambridge, UK.Optical Tweezers for Genomics
Scientists in the US have illuminated the state-of-the-art in optical tweezer research, using laser beams to trap and move small biological objects.Piero Bianco and Yuji Kimura from the University at Buffalo, highlight how researchers can now use optical tweezers to study interactions between DNA and proteins.
Optical tweezers, or optical traps, are conceptually like eyebrow tweezers. An infrared laser is focused through an optical microscope on a precise spot, trapping objects as small as several nanometres across. Moving the laser beam allows these objects to be picked up and moved.
Numerous DNA binding proteins have been studied using optical tweezers (e.g.Fig. 6), including members of the polymerase family. Polymerases are responsible for replicating DNA and transcribing DNA into RNA. Scientists have used optical tweezers to show how a polymerase pauses and back tracks during transcription. This is consistent with a proofreading process, said Bianco. Ulf Landegren from Uppsala University, Sweden, said that the research is proving ‘fundamental for understanding how the genome is regulated.’
 | ||
| Fig. 6 An example of the use of optical tweezers. Reproduced from the Analyst, 2006, 131, 868–874 by permission of The Royal Society of Chemistry. | ||
Optical tweezers have significantly contributed to the understanding of the dynamic behaviour of individual protein molecules at single molecule levels, said Bianco.
He predicts that the method will eventually help scientists to discover how breaks in double stranded DNA are repaired.
Y. Kimura and P. R. Bianco, Analyst, 2006, 131, 868–874.
Reviewed by: Nina Athey-Pollard, Royal Society of Chemistry, Cambridge, UK.Relaxing times for DNA bases
UV-induced skin cancers have a known link to DNA damage. Now scientists in France say the damage can be better understood by looking at how DNA components relax after exposure to light.DNA bases are the only DNA components that can be electronically excited by the sun’s UV radiation, said Clélia Canuel at the Commissariat à l'énergie atomique (CEA) Saclay. Using a technique called time-resolved spectroscopy, Canuel and colleagues have worked out how a derivative of the DNA base adenine loses the energy it absorbs when exposed to UV light.
For the adenine derivative, ‘we found an ultra-fast two step relaxation mechanism,’ said Canuel. This ‘probably prevents chemical reactions in the excited states and mutation introduction,’ continued Canuel, meaning that the adenine is quite stable when exposed to UV light.
Clusters of the adenine derivative and water were also studied, to bridge the gap between isolated molecules and biological environments, explained Canuel. The team found that the base remained stable even when interacting with water.
The researchers will now use the same methods to study the other DNA bases affected by UV radiation: guanine, cytosine and thymine. They plan to examine the bases in isolation and in water clusters, before looking at adenine–thymine and guanine–cytosine pairs.
C. Canuel, M. Elhanine, M. Mons, F. Piuzzi, B. Tardivel and I. Dimicoli, Phys. Chem. Chem. Phys., 2006, 8, 3978–3987.
Reviewed by: Susan Batten, Royal Society of Chemistry, Cambridge, UK.A quick fix for damaged genes
Chemists in Japan are a step closer to repairing damaged DNA inside living cells.Kenzo Fujimoto at the Japan Advanced Institute of Science and Technology and colleagues have developed a technique to target DNA bases. Using their method the researchers can mutate a cytosine base into uracil at a specific site on the DNA backbone. The method is so precise, said Fujimoto, it can select between two cytosine bases adjacent on the DNA strand.
Fujimoto’s method uses a strand of synthetic DNA fitted with a photoreactive end group. The synthetic DNA has a sequence complementary to the DNA targeted for mutation so that the two strands bind together when mixed. UV light triggers a reaction between the photoreactive group and the targeted cytosine and heating converts the tagged cytosine into uracil. The researchers release the selectively mutated DNA using UV irradiation at a different wavelength to break the two strands apart (Fig. 7).
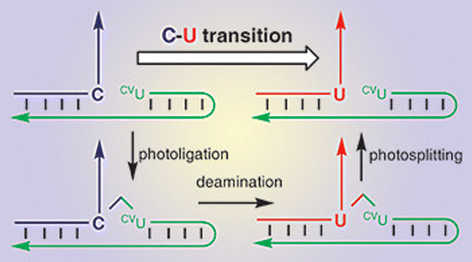 | ||
| Fig. 7 Reproduced from Chem. Comm., 2006, 3223–3225 by permission of the Royal Society of Chemistry. | ||
Although the protocol is currently limited to cytosine, it uses only simple procedures to trigger the mutation, said Fujimoto. Unlike most existing methods the technique does not involve enzymes, and the highly active photoreactive groups mean further UV-active compounds are not needed to bring about the reaction. Because these extra additives are not required, the method could be developed for medical applications, said Fujimoto. For example, ‘the site-specific photochemical DNA manipulation would be used as a tool for photochemical repair [of] damaged DNA.’
Fujimoto’s work has been welcomed by Jerry Davies, an expert in DNA structure and function at Queen’s University, Belfast. ‘While the method does not lend itself directly to introducing site-specific changes at bases other than cytosine, it does provide a successful model that should stimulate further research towards this end,’ said Davies.
K. Fujimoto, S. Matsuda, Y. Yoshimura, T. Matsumura, M. Hayashi and I. Saito, Chem. Comm., 2006, 3223–3225.
Reviewed by: James Mitchell Crow, Royal Society of Chemistry, Cambridge, UK.| This journal is © The Royal Society of Chemistry 2006 |
