Quantitative assessment of olfactory receptors activity in immobilized nanosomes: a novel concept for bioelectronic nose
Jasmina Minic
Vidic
*a,
Jeanne
Grosclaude
b,
Marie-Annick
Persuy
a,
Josiane
Aioun
a,
Roland
Salesse
a and
Edith
Pajot-Augy
*a
aUnité de Neurobiologie de l'Olfaction et de la Prise Alimentaire, Equipe Récepteurs et Communication Chimique, 78352 Jouy-en-Josas Cedex, France. E-mail: jasmina.minic@jouy.inra.fr; marie-annick.persuy@jouy.inra.fr; josiane.aioun@jouy.inra.fr; roland.salesse@jouy.inra.fr; edith.pajot@jouy.inra.fr; Fax: + 33 (0) 1 34 65 22 41; Tel: + 33 (0) 1 34 65 25 63
bUnité de Virologie et Immunologie Moléculaires, INRA, 78352 Jouy-en-Josas Cedex, France. E-mail: jeanne.grosclaude@jouy.inra.fr
First published on 31st May 2006
Abstract
We describe how mammalian olfactory receptors (ORs) could be used as sensing elements of highly specific and sensitive bioelectronic noses. An OR and an appropriate Gα protein were co-expressed in Saccharomyces cerevisiae cells from which membrane nanosomes were prepared, and immobilized on a sensor chip. By Surface Plasmon Resonance, we were able to quantitatively evaluate OR stimulation by an odorant, and G protein activation. We demonstrate that ORs in nanosomes discriminate between odorant ligands and unrelated odorants, as in whole cells. This assay also provides the possibility for quantitative assessment of the coupling efficiency of the OR with different Gα subunits, without the interference of the cellular transduction pathway. Our findings will be useful to develop a new generation of electronic noses for detection and discrimination of volatile compounds, particularly amenable to micro- and nano-sensor formats.
Introduction
Odorants can constitute a signature of metabolic states or diseases, participate in aromas in food, be associated with drugs and explosives or to domestic and environmental pollutants. Therefore, there is an increasing interest in emerging technologies allowing rapid and noninvasive assessment of volatile odorant compounds. Current electronic nose devices based on metal oxide semiconductors or conducting polymers that specifically identify gaseous odorants are typically large and expensive and thus not adequate for use in micro- or nano-arrays that could mimic the performance of the natural olfactory system.1–5 Moreover, they are hampered by the presence of water, they do not involve molecular recognition of odorants, and they require a large sample size to ensure an optimal interaction with sensor surface.2,4,6 Replacing these physical or chemical sensing elements with olfactory receptors (ORs) would provide a new platform with the capacity to overcome these weaknesses, resulting in bioelectronic olfaction devices.The vertebrate olfactory system can distinguish among hundreds of thousands of different volatile molecules exhibiting diverse functional groups, branching patterns, charges or unsaturated bonds, size and shape. The ORs mediating the perception of odorants belong to the large super-family of G protein coupled receptors (GPCRs).7 Each OR can respond to a range of odorants that share specific molecular features, and can discriminate slight discrepancies in odorant structure, size or concentration.8–13 Moreover, an OR can detect its preferential odorant ligand at a very low concentration, i.e. 10 pM–10 nM.13,14
Until now, response specificities to odorants have been examined in detail only for a few ORs. The main reason is that such studies cannot be undertaken in olfactory epithelium since individual sensory neurons mainly express a single type of OR out of several hundred OR genes in the genome.11,15 In addition, there are inherent difficulties associated with the expression of ORs in heterologous cell lines.16,17 In consequence, most assays of OR function have been performed in whole cells or transgenic animals, neither of which complies with a requirement for high throughput screening.
In the present study, we show that ORs expressed in S. cerevisiae retain their full activity in isolated nanosomes immobilized on sensorchips. We characterize the selectivity and sensitivity of the ORs upon odorant detection using Surface Plasmon Resonance (SPR). In addition, we report experiments demonstrating that our approach allows the study of OR coupling efficiency with different Gα subunits, since it discards the possible contribution of the cellular signaling cascade in response to receptor stimulation.
Experimental
Yeast transformations and nanosomes preparation
Yeast strain MC18 (MATa gpa1 :: lacZ [LEU2]ade2-1 his3-11, 15 leu2- 3,112 trp1-1 ura3-1 can1-100), kindly provided by Prof. I. Connerton (University of Nottingham, UK) was transformed using the lithium acetate method.18 Plasmids pJH2-I7, encoding rat ORI7 (ORL11 in OrDB) and pJH2-cmycOR1740, encoding human OR1740 (ORL520 in OrDB) were obtained by homologous recombination in the pJH2-SSTR2 plasmid (a gift from Dr M.H. Pausch, Cyanamid Agricultural Research Center, Princeton, NJ), as described previously.19,20 Plasmids pRGP-Gαolf, pRGP-Gα15 were gifts from Prof. I. Connerton.21 pLP82 encoding chimeric GPA1-Gαi2 protein was kindly provided by M.H. Pausch. Plasmid pLP85 encoding chimeric GPA1-Gαolf protein was obtained as described previously.20Yeast colonies were grown in 0.67% yeast nitrogen base (Difco, Le Pont de Claix, France), synthetic drop-out CSM media without HIS, LEU, TRP, URA (Bio101, Inc., CA, USA), 40 mg ml−1 adenine, complemented with 2% glucose. Expression of ORs was obtained by induction of the transformed yeasts with 2% galactose at 15 °C for 60 h as described previously.20
Membrane fraction was prepared starting from yeast cells washed two times with ice-cold water, harvested by centrifugation and resuspended in an equal volume of ice-cold lysis buffer (50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 0.1 mM PMSF and the Complete protease inhibitor cocktail (Roche, Meylan, France). Glass beads (425–600 µm, Sigma-Aldrich, Saint Quentin Fallavier, France) were added and cells were disrupted by 7 cycles of 1 min vigorous vortexing/1 min of cooling on ice. Samples were pooled and centrifuged at 5000 g for 10 min at 4 °C to remove unbroken cells and cell wall fragments. The supernatant was then ultra-centrifuged at 40,000 g for 40 min at 4 °C. This pellet, enriched in membrane fraction, was resuspended in the lysis buffer and 10% (w/w) glycerol with a Dounce homogenizer, and stored in aliquots at −80 °C. The protein concentration of the membrane preparation was determined using the BCA reagent (Pierce, Brebieres) with bovine serum albumin as a standard. Membrane fraction can be prepared in large amounts, frozen and stored for many weeks without losing biological activity.
Immediately before use, the membrane fraction was sonicated for 20 min at 120 W, 42 kHz (Deltasonic sonicator, Meaux, France) in ice-cold water.
Antibodies
The mouse anti-cmyc monoclonal antibody was from Roche Molecular Biochemical. The rabbit anti-I7 polyclonal antibody raised against its N-terminal 15 aminoacids was custom-made by Neosystem (Strasbourg, France). The anti-rabbit F(ab′)2 fragments antibody coupled to 10 nm gold beads was from Aurion (The Netherlands).Odorants
Odorant solutions were prepared just before use as described previously.13,20 Octanal, nonanal, heptanal, diacetyl, cyclohexyl-acetate, octanol, octanone, octanoic acid, isoamyl-acetate, pyridine were from Sigma-Aldrich. Helional, cassione, piperonyl acetate, 3,4-methylenedioxyphenyl acetone and 3,4-methylenedioxypropiophenone were generous gifts from Givaudan-Roure (Switzerland), courtesy of Boris Schilling. Lyral and lilial were kindly provided by Roche.Electron microscopy
Yeast cell fixation and immunostaining procedures were performed as described previously.19 Visualization of microsomes and nanosomes by negative staining was carried out as described in ref. 22 under a CM12 Philips electron microscope.SPR experiments
SPR experiments were conducted on a BIAcore 3000 (Pharmacia Biosensor, Sweden). Nanosomes were diluted in HBS buffer (25 mM HEPES, pH 7.4, containing 150 mM NaCl) before deposition. Protein solutions were run on a L1 chip surface at a low flow rate (2 µl min−1) for 20 min until a stable resonance unit (RU) level was obtained, indicating that binding had reached saturation. Protein-free buffer solution was then injected to rinse-off non-captured material and reach deposition equilibrium. Functional tests were performed with odorants at various concentrations and 10 µM GTP (or GTPγS) in HBS, at a 5 µl min−1 flow rate. In control experiments, stimulation was carried out using solutions in which the odorant had been replaced by water. The biosensor chip was washed several times with 20 mM CHAPS between experiments for regeneration.All measurements were performed at 25 °C. RU levels were compared before and after each injection, to evaluate the amount of material grafted onto the sensor chip.
Results and discussion
Nanosomes preparation and sensor coating
In this study, ORs and Gα proteins were co-expressed in the S. cerevisiae strain MC18.21 This strain was chosen because plasma membrane insertion of a functional OR has previously been shown to be highly efficient.19 Furthermore, the hijacking of yeast pheromone response pathway to induce luciferase reporter synthesis upon receptor stimulation allows us to perform functional tests on the ORs, in order to characterize them before being used as biosensor sensing elements. This yeast strain was transformed to co-express either full length rat ORI7, or human OR 1740 tagged with c-myc sequence on its N-terminus, and the cognate Gαolf subunit. These ORs were chosen as model receptors to investigate the OR functional immobilization onto the sensor chip surface, since their preferential ligands and working concentration ranges are known.19,23,24Yeast cells were mechanically disrupted, as described in the Experimental section, and the membrane fraction was isolated in the form of microsomes with sizes ranging from 40 to 700 nm. Additional sonication was performed on these vesicles to homogenize them and reduce their sizes. Consequently, nanosomes with diameters close to 50 nm were obtained, as visualized by negative staining electron microscopy (Fig. 1a). The nanosomes were also characterized by means of Atomic Force Microscopy and by ultrastructural visualization of samples prepared by cryo-fracture (data to be published), which confirmed the size and size-distribution of sonicated and non-sonicated nanosomes.
 | ||
| Fig. 1 Preparation of yeast nanosomes and their immobilization on SPR sensor chips. (a) A yeast strain transformed with pJH2-I7 plasmid was immuno-labeled with the primary anti-I7 antibody and the 10 nm gold-conjugated secondary antibody. Arrows indicate gold grains present on the plasma membrane of yeast cells (left picture). After cell mechanical disruption, the membrane fraction was isolated by ultracentrifugation in the form of lipid microsomes (diameters 40 to 700 nm, middle picture). The membrane fraction in the form of nanosomes with diameters close to 50 nm was obtained by additional sonication of microsomes. Visualization of these nanosomes by negative staining electronic microscopy shows that most of them are spherical (o), but some appear as open (▲) or closed collapsed fragments (★) (right picture). (b) Schematic illustration of cmyc-OR1740 immobilization on L1 sensor chip and detection with a specific monoclonal antibody against a cmyc-tag at its N-terminus. (c) Immobilization and detection of OR was monitored by SPR (left panel): (1) Nanosomes diluted at various total protein concentrations in HBS buffer were run onto the sensor surface, then (2) HBS buffer was run to remove unbound membrane fragments. (3) Anti-cmyc antibody was injected, and (4) unbound antibody was rinsed away with a flow of HBS buffer. The amount of deposited nanosomes is a function of the total protein concentration in the deposited nanosomes membrane fraction (middle panel). SPR responses then provide a linear correlation with the amount of deposited nanosomes (right panel), giving evidence of the SPR detection of immobilized membrane-bound receptor by its antibody. The SPR signal is expressed as arbitrary resonance units (RU). | ||
The nanosomes were immobilized on a Biacore sensor chip L1 whose surface consists of a covalently linked carboxymethyl-modified dextran polymer hydrogel on which an important part of the glucose moieties are grafted with lipophilic alkyl chains.25 Nanosomes were efficiently hooked by those alkyl anchors, resulting in a very stable deposition yielding a SPR response proportional to the total protein concentration of loaded material. In the case of cmyc-OR1740 nanosomes, the monoclonal anti-cmyc antibody (1 µM) was subsequently run on the sensor chip to detect the receptor present in the immobilized vesicles (Figs. 1b and 1c). The observed SPR response signal was a linear function of the amount of bound nanosomes, suggesting a quantitative detection of cmyc-OR1740. This procedure of ORs preparation and immobilization on a surface offers several advantages over current assay procedures for GPCRs. First, the receptor remains at all times in its native membrane environment, thus obviating the risk of receptor alteration or activity loss, which may occur when GPCRs are reconstituted in artificial membranes or liposomes. Second, since ORs are present at low concentration in the membrane fraction, receptor purification in detergents before reconstitution cannot be considered. Furthermore, ORs are bound on the chip indirectly, through the interaction of their surrounding lipidic bilayer with the substrate, and not directly by binding of their amino-acid chain. This feature warrants that the receptor binding site for odorants remains accessible and functional. Finally, the Gα protein subunit remains anchored to the nanosomes bilayer allowing the detection of receptor activity using a previously evaluated method for monitoring rhodopsin receptor activation by light.26
Activity of olfactory receptors in immobilized nanosomes
As the SPR signal is proportional to the molecular weight of the analyte that binds to the sample on the chip surface, low-molecular-weight compounds, such as the majority of odorants, cannot be detected directly.27 To overcome this problem, we took advantage of the presence of Gαolf anchored to the nanosomes to monitor receptor activation by an odorant ligand, through the desorption of Gαolf subunit from the lipidic bilayer (Fig. 2a).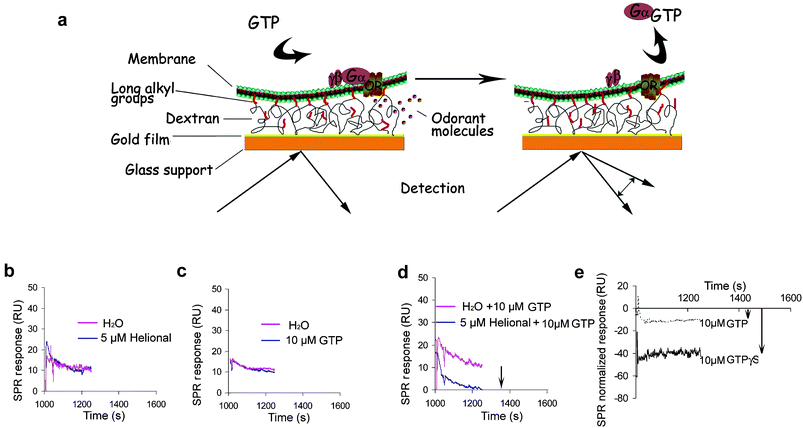 | ||
| Fig. 2 Principle of the sensor system. (a) The L1 sensor chip (BIAcore) is composed of a gold film deposited onto a glass support, on which a carboxylated dextran grafted with long alkyl groups is attached. Nanosomes are immobilized via interaction of their lipidic surface with these long alkyl chains, leaving membrane receptors fully available for ligands and G protein interactions. The scheme presents only a restricted area of the lipidic bilayer, either within an intact nanosome or part of an extended lipidic bilayer obtained from ruptured and fused nanosomes. Upon stimulation of an OR with an odorant ligand, the Gα subunit is activated and desorbed in the presence of GTP. This results in a shift of the SPR response level expressed as arbitrary resonance units (RU). No shift of the SPR response level is observed when nanosomes are stimulated either with odorant alone (b), or GTP alone (c), as compared to the control stimulated with water. The signal modification is only observed when odorant and GTP are injected simultaneously, as compared to the control stimulated with water and GTP (d). The signal relative to the release of the Gα subunit can be enhanced 4-fold by replacing GTP by GTPγS. SPR responses are normalized as the differential relative to the corresponding controls obtained by replacing odorant with water (e). | ||
To assess OR activity, we performed several tests on cmyc-OR1740 immobilized nanosomes. Helional is the preferential ligand of OR1740.13,19,23 As expected, no response was observed when helional alone was run onto the nanosomes (Fig. 2b), and no SPR response was observed either when GTP alone was used, indicating that immobilized ORs are not constitutively active (Fig. 2c). However, an easily measurable modification of SPR signal level was induced by injection of both helional and GTP (Fig. 2d), resulting from the activation of the trimeric G protein and mobilization of Gαolf. The sensitivity of the assay was enhanced about four-fold when GTP was replaced by its non-hydrolysable analogue GTPγS (Fig. 2e). No shift of SPR signal was observed when helional was replaced by an unrelated odorant under the same experimental conditions (see below). Based on these findings, the desorption signal can be considered as a reporter of the functional response of the OR, since the receptor has to be stimulated by a ligand to induce G protein mobilization.
Detection of odorants
We then investigated whether cmyc-OR1740 retains its sensing characteristics in immobilized nanosomes by assessing the responses to different concentrations of various odorants (Fig. 3). Comparing responses of this device with those of the reporter gene transcription method in whole yeast cells expressing the receptor allowed us to acknowledge several key common features. First, cmyc-OR1740 receptor in chip-bound nanosomes and in whole yeast displays helional as the most potent agonist. The specificity of the SPR response was additionally cross-tested by stimulation of another receptor, ORI7, by octanal, one of its ligands, and helional as a negative control (data not shown). In contrast to OR1740, ORI7 cannot be activated by helional, but induces a SPR response when stimulated with octanal. Second, the detection of helional analogues by sensor chip-bound receptor indicates that the odorants are ranked in the same order (helional > cassione > piperonyl acetate, 3,4-methylenedioxyphenyl acetone, 3,4-methylenedixypropiophenone) and on the same molecular receptive range, as in whole cells. Third, no response was observed with unrelated odorants octanal and vanillin. Fourth, we observed the same high sensitivity of the receptor, since SPR curves show the same low agonist thresholds as observed with the bioluminescence reporter in whole yeast cells. Finally, both methods exhibit the same bell-shaped odorant concentration dependence. The two maxima observed for functional response may arise from multiple agonist/antagonist binding sites with different affinities available on a receptor. The molecular mechanisms of functional interactions between odorants and ORs are indeed still a subject of investigation. | ||
| Fig. 3 Detection of odorants by cmyc-OR1740 in immobilized nanosomes and by the receptor in whole yeast cells. (a) Differential SPR responses obtained upon stimulation of the ORs in nanosomes with various odorants in the presence of 10 µM GTPγS. (b) Differential bioluminescence responses upon stimulation of the receptor in whole yeast with the same panel of odorants. In both cases, concentration-dependent curves are plotted as a difference of response to odorants relative to controls obtained by replacing odorants with water. Odorants tested are shown in the middle panel. | ||
Overall, these data demonstrate that receptor activity is not hampered by immobilization of nanosomes, since selectivity and high sensitivity of a mammalian OR are maintained in the device.
Stability of the ORs in immobilized nanosomes
Next, in order to check whether this kind of device could be used in successive trials, the same experiment was carried out several times in a row. A lag phase of 30 min was needed for the system to recover its response ability. Strikingly, the system displayed the same activity level over several activation cycles (up to 8) one hour apart from each other (Fig. 4). No further response was detected thereafter, either due to depletion of the pool of Gαolf bound to the nanosomes, or because a reorganization of the immobilized nanosomes took place at the sensor surface.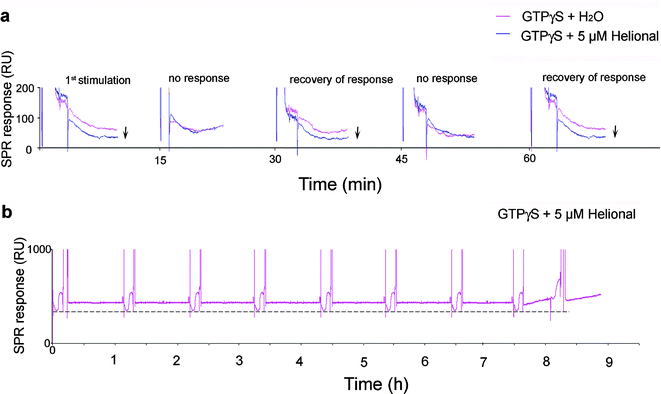 | ||
| Fig. 4 Successive responses of the immobilized cmyc-OR1740 receptor. (a) If the cmyc-OR1740 receptor is stimulated several times in a row with 5 µM helional and 10 µM GTPγS, a minimum 30 min lapse-time is necessary between two stimulations to observe its activity. (b) If a 1 h lapse-time is allowed between two consecutive injections of 5 µM helional and 10 µM GTPγS, the same activity level (- - -) is observed from the receptors in immobilized nanosomes for up to 8 activation cycles. | ||
This stability and repeatability of response obtained with immobilized nanosomes as miniaturized sensing elements is an advantage over whole yeast cells that can only be single-used in functional tests.
OR coupling with a variety of Gα subunits
In order to show that our device allows rapid evaluation of the efficiency of an OR coupling to Gα subunits, nanosomes were prepared from yeast strains co-expressing ORI7 and various Gα subunits, immobilized on the sensor chip and stimulated with receptor ligand octanal (5 µM) and 10 µM GTPγS (Fig. 5). Since the same level of response is measured during several stimulation cycles, we figured that the amount of Gα protein bound to the nanosomes is not a limiting factor for the signal intensity upon initial odorant stimulation.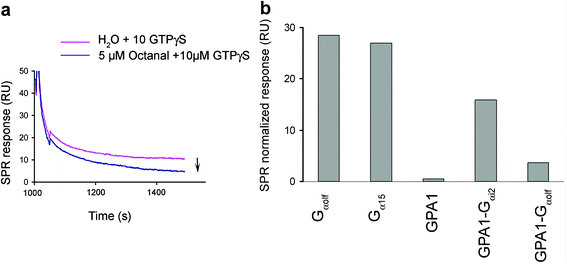 | ||
| Fig. 5 Efficiency of coupling of immobilized ORI7 with different Gα subunits. (a) Nanosomes prepared from ORI7 strain additionally transformed to co-express Gαolf subunit were immobilized on the sensor chip and stimulated with 5 µM octanal and 10 µM GTPγS. Receptor stimulation by the odorant resulted in the decrease of the SPR response as presented in overlay plots for octanal and water stimulations. (b) Relative decrease data were normalized to the amount of nanosomes immobilized. | ||
The maximal intensity of response to octanal was observed with nanosomes bearing ORI7 and its cognate Gαolf protein (Fig. 5a). ORI7 was not able to couple efficiently with yeast endogenous Gα subunit, GPA1, since no desorption signal was observed in nanosomes prepared from this yeast strain. This result correlates to a previous study of ORI7 in whole yeast cells.20 Furthermore, as also observed in these in vivo yeast assays, we give evidence that the coupling is improved with chimeric versions of GPA1, namely GPA1-Gαi2 and GPA1-Gαolf. However, in contrast to the weak coupling efficiency of I7 with Gα15 protein reported in another whole yeast cells assay,19 a highly efficient coupling is observed in the present experiments (Fig. 5b). This corroborates the assumption that Gα15 and its chimeras have a poor affinity for yeast Gβγ subunits, which are indeed involved in the cellular assay28,29 but a high affinity for ORs.10,30,31
Most yeast-based GPCR assays hijack the S. cerevisiae pheromone signaling pathway to activate a reporter gene transcription upon receptor stimulation by a ligand. The advantage of the sensor chip-based assay for evaluation of receptor activity and of its interaction with various Gα subunits is that it obviates cellular signaling pathway interferences. In this respect, our strategy to use nanosomes instead of whole cells may prove useful for the assessment of GPCRs with unknown cognate Gα subunit, or cellular signaling pathway.
Conclusions and perspectives
We have developed and validated a highly specific and sensitive odor-sensing device based on ORs as biosensors. The key feature of the system is that both OR and Gα protein are borne by nanosomes immobilized on the chip surface, thus enabling direct and label-free detection of a functional response. An OR in immobilized nanosomes discriminates between its preferential ligand and a number of analogues, as it does in whole cells, which substantiates the fact that its activity is preserved in these operating conditions devoid of living cells. Working with ORs in nanosomes allows eliminating the possible contribution of the cellular transduction pathway to the response arising from the receptor itself upon odorant stimulation. Such a sensor also enables comparison of the interaction efficiency of ORs with different Gα subunits, and may prove useful for investigation of peripheral mechanisms for odorant detection and coding.This chip assay gains benefit from using yeast cells as a host for OR expression, such as low cost, simplicity for genetic manipulations, a “null background” due to the lack of endogenous GPCRs.32 Moreover, it exhibits definite advantages as compared to commonly used yeast-based assays that couple a GPCR to the pheromone signaling pathway. First, the yeast cell wall restriction for ligand penetration is lifted, and our assay can be employed for direct investigation of GPCRs with high molecular weight ligands. Second, the functional characterization of many GPCRs expressed in yeast is usually possible due to their coupling to chimeric yeast-mammalian Gα proteins.29 Therefore, it is usually difficult to ascertain the physiological coupling specificity of a receptor. Here we showed that such investigations can indeed be carried out on receptors borne by nanosomes prepared from genetically modified yeasts.
This chip assay format requires only minute quantities of nanosomes suggesting that our sensor design has a potential for micro- and nano-scale sensor applications. In addition, the SPR method described here is suitable for high throughput screening of GPCRs because immobilized receptors can be stimulated repetitively without any special handling, just allowing a minimum 30 min lapse-time between two successive stimulations, and methods for automatic SPR analysis are now well established.33
The development of sensor technology incorporating natural ORs would provide the basis for a bioelectronic nose mimicking the vertebrate olfactory system. Such a device could be used to identify and monitor a spectrum of odorants in real time with a much higher selectivity and specificity than present electronic devices. Current research in our group focuses on the development of a nanobiosensor array based on immobilized nanosomes carrying ORs of interest for label-free electrical detection of odorants (SPOT-NOSED European Project).
Acknowledgements
We thank Prof. I. Connerton (University of Nottingham) and Dr M. Pausch (Cyanamid Agricultural Research Center) for providing the yeast strain and plasmids used in this study, Drs B. Schilling and S. Bieri (Givaudan-Roure) for the gift of helional and its analogues. We also wish to warmly thank D. Grebert and R. Monnerie for their dedicated and skillful support and all members of NOPA-RCC and SPOT-NOSED consortium for critical comments and discussion. This work was financially supported by the SPOT-NOSED Project of the European Community (IST-2001-38739), the Ile-de-France region, in the framework of a SESAME contract (2002/AO1497), and the PICASSO program (HF2004-0055) funded by EGIDE.References
- T. A. Dickinson, J. White, J. S. Kauer and D. R. Walt, Current trends in ‘artificial-nose’ technology, Trends Biotechnol., 1998, 16(6), 250–8 CrossRef CAS.
- A. P. Turner and N. Magan, Electronic noses and disease diagnostics, Nat. Rev. Microbiol., 2004, 2(2), 161–6 Search PubMed.
- K. Persaud and G. Dodd, Analysis of discrimination mechanisms in the mammalian olfactory system using a model nose, Nature, 1982, 299(5881), 352–5 CAS.
- W. J. Harper, The strengths and weaknesses of the electronic nose, Kluwer Academic/Plenum Publishers, New York, 2001 Search PubMed.
- E. Vanneste and H. J. Geise, Commercial electronic nose instruments, WILEY-VCH, Weinheim, 2003 Search PubMed.
- T. C. Pearce, S. S. Schiffman, H. T. Nagle and J. W. Gardner, Handbook of Machine Olfaction: Electronic Nose Technology, Wiley, 2002 Search PubMed.
- L. Buck and R. Axel, A novel multigene family may encode odorant receptors: a molecular basis for odor recognition, Cell, 1991, 65(1), 175–87 CAS.
- R. C. Araneda, A. D. Kini and S. Firestein, The molecular receptive range of an odorant receptor, Nat. Neurosci., 2000, 3(12), 1248–55 CrossRef CAS.
- B. Malnic, J. Hirono, T. Sato and L. B. Buck, Combinatorial receptor codes for odors, Cell, 1999, 96(5), 713–23 CrossRef CAS.
- K. Kajiya, K. Inaki, M. Tanaka, T. Haga, H. Kataoka and K. Touhara, Molecular bases of odor discrimination: Reconstitution of olfactory receptors that recognize overlapping sets of odorants, J. Neurosci., 2001, 21(16), 6018–25 CAS.
- P. Mombaerts, Genes and ligands for odorant, vomeronasal and taste receptors, Nat. Rev. Neurosci., 2004, 5(4), 263–78 CrossRef CAS.
- H. Breer, Olfactory receptors: molecular basis for recognition and discrimination of odors, Anal. Bioanal. Chem., 2003, 377(3), 427–33 CrossRef CAS.
- G. Levasseur, M. A. Persuy, D. Grebert, J. J. Remy, R. Salesse and E. Pajot-Augy, Ligand-specific dose-response of heterologously expressed olfactory receptors, Eur. J. Biochem., 2003, 270(13), 2905–12 CrossRef CAS.
- R. C. Araneda, Z. Peterlin, X. Zhang, A. Chesler and S. Firestein, A pharmacological profile of the aldehyde receptor repertoire in rat olfactory epithelium, J. Physiol., 2004, 555(Pt 3), 743–56 CAS.
- P. Mombaerts, Odorant receptor gene choice in olfactory sensory neurons: the one receptor-one neuron hypothesis revisited, Curr. Opin. Neurobiol., 2004, 14(1), 31–6 CrossRef CAS.
- M. Lu, F. Echeverri and B. D. Moyer, Endoplasmic reticulum retention, degradation, and aggregation of olfactory G-protein coupled receptors, Traffic, 2003, 4(6), 416–33 CrossRef CAS.
- T. S. McClintock and N. Sammeta, Trafficking prerogatives of olfactory receptors, NeuroReport, 2003, 14(12), 1547–52 Search PubMed.
- R. H. Schiestl, P. Manivasakam, R. A. Woods and R. D. Gietz, Introducing DNA into yeast by transformation, Methods, 1993, 5, 79–85 Search PubMed.
- J. Minic, M. A. Persuy, E. Godel, J. Aioun, I. Connerton, R. Salesse and E. Pajot-Augy, Functional expression of olfactory receptors in yeast and development of a bioassay for odorant screening, FEBS. J., 2005, 272(2), 524–37 Search PubMed.
- E. Pajot-Augy, M. Crowe, G. Levasseur, R. Salesse and I. Connerton, Engineered yeasts as reporter systems for odorant detection, J. Recept. Signal Transduct. Res., 2006, 23(2–3), 155–71.
- M. L. Crowe, B. N. Perry and I. F. Connerton, Golf complements a GPA1 null mutation in Saccharomyces cerevisiae and functionally couples to the STE2 pheromone receptor, J. Recept. Signal Transduct. Res., 2000, 20(1), 61–73 CrossRef CAS.
- J. Minic, J. Grosclaude, J. Aioun, M. A. Persuy, T. Gorojankina, R. Salesse, E. Pajot-Augy, Y. Hou, S. Helali, N. Jaffrezic-Renault, F. Bessueille, A. Errachid, G. Gomila, O. Ruiz and J. Samitier, Immobilization of native membrane-bound rhodopsin on biosensor surfaces, Biochim. Biophys. Acta, 2005, 1724(3), 324–32 CrossRef CAS.
- C. H. Wetzel, M. Oles, C. Wellerdieck, M. Kuczkowiak, G. Gisselmann and H. Hatt, Specificity and sensitivity of a human olfactory receptor functionally expressed in human embryonic kidney 293 cells and Xenopus Laevis oocytes, J. Neurosci., 1999, 19(17), 7426–33 CAS.
- H. Zhao, L. Ivic, J. M. Otaki, M. Hashimoto, K. Mikoshiba and S. Firestein, Functional expression of a mammalian odorant receptor, Science, 1998, 279(5348), 237–42 CrossRef CAS.
- H. Mozsolits, W. G. Thomas and M. I. Aguilar, Surface plasmon resonance spectroscopy in the study of membrane-mediated cell signalling, J. Pept. Sci., 2003, 9(2), 77–89 CrossRef CAS.
- C. Bieri, O. P. Ernst, S. Heyse, K. P. Hofmann and H. Vogel, Micropatterned immobilization of a G protein-coupled receptor and direct detection of G protein activation, Nat. Biotechnol., 1999, 17(11), 1105–8 CrossRef CAS.
- R. Karlsson, Real-time competitive kinetic analysis of interactions between low-molecular-weight ligands in solution and surface-immobilized receptors, Anal. Biochem., 1994, 221(1), 142–51 CrossRef CAS.
- S. J. Dowell and A. J. Brown, Yeast assays for G-protein-coupled receptors, Recept. Channels, 2002, 8(5–6), 343–52 Search PubMed.
- A. J. Brown, S. L. Dyos, M. S. Whiteway, J. H. White, M. A. Watson, M. Marzioch, J. J. Clare, D. J. Cousens, C. Paddon, C. Plumpton, M. A. Romanos and S. J. Dowell, Functional coupling of mammalian receptors to the yeast mating pathway using novel yeast/mammalian G protein alpha-subunit chimeras, Yeast, 2000, 16(1), 11–22 CrossRef CAS.
- E. Shirokova, K. Schmiedeberg, P. Bedner, H. Niessen, K. Willecke, J. D. Raguse, W. Meyerhof and D. Krautwurst, Identification of specific ligands for orphan olfactory receptors. G protein-dependent agonism and antagonism of odorants, J. Biol. Chem., 2005, 280(12), 11807–15 CrossRef CAS.
- D. Krautwurst, K. W. Yau and R. R. Reed, Identification of ligands for olfactory receptors by functional expression of a receptor library, Cell, 1998, 95(7), 917–26 CrossRef CAS.
- J. Minic, M. Sautel, R. Salesse and E. Pajot-Augy, Yeast system as a screening tool for pharmacological assessment of g protein coupled receptors, Curr. Med. Chem., 2005, 12(8), 961–9 CrossRef CAS.
- M. Fivash, E. M. Towler and R. J. Fisher, BIAcore for macromolecular interaction, Curr. Opin. Biotechnol., 1998, 9(1), 97–101 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2006 |
