Ring-opening polymerization of coordination complexes: silver(I) complexes with bis(amidopyridine) ligands derived from thiophene
Nancy L. S. Yue, Michael C. Jennings and Richard J. Puddephatt*
Department of Chemistry, University of Western Ontario, London, Canada N6A 5B7. E-mail: pudd@uwo.ca
First published on 23rd June 2006
Abstract
The thiophene-based bis(N-methylamido-pyridine) ligand SC4H2-2,5-{C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)N(Me)-4-C5H4N}2 reacts with silver(I) salts AgX to give 1 : 1 complexes, which are characterized in the solid state as the macrocyclic complexes [Ag2{SC4H2-2,5-(CONMe-4-C5H4N)2}2][X]2, which have the cis conformation of the C(
O)N(Me)-4-C5H4N}2 reacts with silver(I) salts AgX to give 1 : 1 complexes, which are characterized in the solid state as the macrocyclic complexes [Ag2{SC4H2-2,5-(CONMe-4-C5H4N)2}2][X]2, which have the cis conformation of the C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)N(Me) group, when X = CF3CO2, NO3, or CF3SO3 but as the polymeric complex [Agn{SC4H2-2,5-(CONMe-4-C5H4N)2}n][X]n, with the unusual trans conformation of the C(
O)N(Me) group, when X = CF3CO2, NO3, or CF3SO3 but as the polymeric complex [Agn{SC4H2-2,5-(CONMe-4-C5H4N)2}n][X]n, with the unusual trans conformation of the C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)N(Me) group, when X = PF6. The bis(amido-pyridine) ligand SC4H2-2,5-{C(
O)N(Me) group, when X = PF6. The bis(amido-pyridine) ligand SC4H2-2,5-{C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)NHCH2-3-C5H4N}2 reacts with silver(I) trifluoroacetate to give the polymeric complex [Agn{SC4H2-2,5-(CONHCH2-3-C5H4N)2}n][X]n, X = CF3CO2. The macrocyclic complexes contain transannular argentophilic secondary bonds. The polymers self assemble into sheet structures through interchain C
O)NHCH2-3-C5H4N}2 reacts with silver(I) trifluoroacetate to give the polymeric complex [Agn{SC4H2-2,5-(CONHCH2-3-C5H4N)2}n][X]n, X = CF3CO2. The macrocyclic complexes contain transannular argentophilic secondary bonds. The polymers self assemble into sheet structures through interchain C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O⋯Ag and S⋯Ag bonds in [Agn{SC4H2-2,5-(CONMe-4-C5H4N)2}n][PF6]n and through Ag⋯Ag, C
O⋯Ag and S⋯Ag bonds in [Agn{SC4H2-2,5-(CONMe-4-C5H4N)2}n][PF6]n and through Ag⋯Ag, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O⋯Ag and Ag⋯O(trifluoroacetate)⋯HN secondary bonds in [Agn{SC4H2-2,5-(CONHCH2-3-C5H4N)2}n][CF3CO2]n.
O⋯Ag and Ag⋯O(trifluoroacetate)⋯HN secondary bonds in [Agn{SC4H2-2,5-(CONHCH2-3-C5H4N)2}n][CF3CO2]n.
Introduction
There is continuing interest in ring-opening polymerization (ROP) of macrocyclic transition metal complexes to give polymers with metals in the backbone structure.1 In labile complexes, rapid reversible ring-opening of macrocyclic complexes can occur in solution, and crystallization can then yield either the ring or polymer form.1,2 The preferred form in solution may be determined by template effects involving guest or solvent molecules, or by secondary bonding involving counterions. The preferred form in the crystalline state may also depend on supramolecular interactions,1–3 and there are a few cases in which both ring and polymer forms are present in the same crystal.4,5 Silver(I) has been used extensively in the self-assembly of polymers and network materials because it forms substitutionally labile complexes and can easily adopt different stereochemistries, with coordination numbers ranging from two to four.6In previous papers, it has been shown that flexible bidentate or tridentate ligands containing two N-methyl-amido-4-pyridine arms, 1 and 2 (Chart 1), can give either macrocyclic disilver(I) complexes, 3 and 4 (Chart 1), or a polymeric complex, 5 (Chart 1), in the solid state.5,7 The complexes with X = CH (Chart 1) formed only macrocyclic complexes 3 in which the ligands adopted the distorted cis,cis,syn conformation, and the cavity of the macrocycle could expand to include a guest anion or contract to form a transannular Ag⋯Ag secondary bond.6 When X = N (Chart 1), the complexes could crystallize either as macrocycles 4 or as a mixture of macrocycle 4 and polymer 5, and the extra pyridyl group could coordinate to an extra silver(I) ion or increase the dimensionality through intermolecular N⋯Ag bonding.5
 | ||
| Chart 1 | ||
In this paper, the chemistry of the analogous ligand with a central thiophene group in place of the benzene or pyridine group of 1 or 2 (Chart 1) is described. The ligand 6 (Chart 2) can adopt either the cis,cis,syn conformation 6-A or the trans,trans,syn conformation 6-B in forming silver(I) complexes. The thiophene group of 6 has the potential to act as a weak ligand for silver(I). In general, thiophene is a weaker ligand than the central pyridine group present in ligand 2, but perhaps a stronger ligand than the central phenyl group (with the potential ability to form arene complexes) of ligand 1. Hence the thiophene groups could increase the dimensionality of the silver(I) compounds, whose primary structures are formed by bonding between silver(I) and the pyridyl arms of ligand 6. The ligand 7 contains both a central thiophene group and simple amide units, so it could give supramolecular chemistry through either hydrogen bond formation or secondary bonding using the thiophene donor.8 By studying this ligand, it was hoped to determine the relative importance of the two types of secondary bonding. Of course, all the complexes also have the potential to undergo supramolecular association through argentophilic bonding.9 The use of thiophene groups in supramolecular chemistry has been explored previously, and it is well known that electropolymerized thiophene derivatives can coordinate to silver(I).10,11
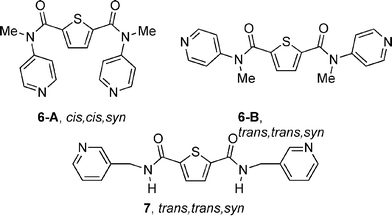 | ||
| Chart 2 | ||
Results
The ligand SC4H2-2,5-{C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/h3_char_e001.gif) O)N(Me)-4-C5H4N}2, 6
O)N(Me)-4-C5H4N}2, 6
The structure of the protonated form of ligand 6, studied as 6·2HCl·4H2O, is shown in Fig. 1, and some conformational parameters are listed in Table 1. There is a mirror plane which bisects the thiophene unit, and the ligand adopts a distorted form of the cis,cis,syn conformation. The chloride anions are hydrogen bonded to the pyridinium NH groups, with distance N(11)⋯Cl(24) = 2.961(5) Å. There is a water molecule, which lies on the mirror plane, and is hydrogen bonded to the chloride anions with distance O(5A)⋯Cl(24) = 2.923(5) Å, and several other water molecules that are not shown. The thiophene unit is twisted out of the amide plane with the sulfur atom syn to the carbonyl oxygen atom and so directed away from the pyridyl groups (Table 1, Fig. 1).
| N⋯Na | Ag⋯Aga | Θ1b | Θ2c | Θ3d | |
|---|---|---|---|---|---|
| a Distance between pyridyl nitrogen atoms or between silver(I) centers in Å.b Θ1 = Dihedral angle SCCO, a measure of the twist from the syn-planar conformation of the thiophene with respect to carbonyl group of the amide unit.c Θ2 = Dihedral angle CCNC(py or CH2py), twist of the amide unit from the cis-planar conformation.d Θ3 = Dihedral angle C(carbonyl)NCC(py or CH2py), twist of the pyridyl or pyridylmethyl group with respect to the amide unit. | |||||
| 6·2HCl·4H2O | 5.65 | 47 | 23 | 30 | |
| 8a | 4.06 | 3.33 | 147, 144 | 30, 26 | 33, 38 |
| 3.66 | 3.06 | 135, 142 | 30, 33 | 42, 38 | |
| 8b | 3.58 | 3.14 | 142, 132 | 32, 23 | 34, 47 |
| 3.50 | 3.04 | 149, 142 | 30, 28 | 45, 40 | |
| 8c | 3.88 | 3.42 | 146, 140 | 28, 32 | 42,39 |
| 4.10 | 3.60 | 146, 134 | 36, 25 | 38, 33 | |
| 8d | 15.53 | 19.78 | 31, 41 | 168, 165 | 5, 22 |
| 7 | 13.1 | 18, 7 | 178, 170 | 85, 78 | |
| 9 | 14.1 | 18.2 | 23, 12 | 175, 180 | 96, 88 |
 | ||
| Fig. 1 A view of the structure of the protonated ligand C4H2S(CONMe-4-C5H4N)2, 6, in 6·2HCl·4H2O. The water molecules which are not hydrogen bonded to either 6 or the Cl− ions are not shown. Symmetry equivalent: A, x, 1.5 − y, z. | ||
Silver(I) complexes with ligand 6
Reaction of silver(I) salts AgX with the ligand 6 gave the corresponding complexes of stoichiometry [{Ag(µ-6)}n]Xn, 8a, X = CF3CO2; 8b, X = NO3; 8c, X![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CF3SO3; 8d, X = PF6. The products can be isolated in crystalline form as either macrocycles (n = 2, 8a–8c) or as polymer (n = 4, 8d), whose structures are illustrated in Scheme 1 and described below.
CF3SO3; 8d, X = PF6. The products can be isolated in crystalline form as either macrocycles (n = 2, 8a–8c) or as polymer (n = 4, 8d), whose structures are illustrated in Scheme 1 and described below. | ||
| Scheme 1 | ||
The structure of the trifluoroacetate derivative 8a, is shown in Fig. 2, with conformational data listed in Table 1. The complex exists as the disilver(I) macrocycle in an extended chair conformation. There are two independent, similar molecules, each of which contains an inversion centre, and only one is shown in Fig. 2. Each silver atom is bound to two pyridyl nitrogen atoms and, more weakly, to two asymmetrically bridging κ1-µ2-trifluoroacetate ions, which are situated above and below the macrocycle (Fig. 2). There is also a transannular argentophilic interaction [molecule 1, Ag(1)⋯Ag(1A) = 3.3266(6); molecule 2, Ag(2)⋯Ag(2B) = 3.0604(7) Å]. The macrocycles are arranged in chains but the intermolecular silver⋯silver distance of Ag(1)⋯Ag(2B) = 3.55 Å is too long to represent a significant bonding interaction.9 Compared to the protonated ligand, the thiophene group is rotated with respect to the amide unit so that the sulfur is directed towards the pyridyl groups, and this allows a shorter N⋯N separation between the pyridine donor atoms (Table 1, Fig. 1 and 2).
![A view of the structure of the trifluoroacetate complex [Ag2(CF3CO2)2{C4H2S(CONMe-4-C5H4N)2}2], 8a. Selected bond parameters: Ag(1)–N(11) 2.195(2); Ag(1)–N(31) 2.182(2); Ag(1)–O(81) 2.522(2); Ag(1)–O(81A) 2.655(3); Ag(1)–Ag(1A) 3.3265(6) Å; N(31)–Ag(1)–N(11) 160.4(1); N(31)–Ag(1)–O(81) 104.9(1); N(11)–Ag(1)–O(81) 89.6(1)°. Symmetry equivalents: A, −x, 2 −
y, −2 −
z. For the independent molecule, corresponding parameters are: Ag(2)–N(21) 2.145(2); Ag(2)–N(41) 2.160(3); Ag(2)–O(71) 2.500(3); Ag(2)–O(71A) 2.677(3); Ag(2)–Ag(2A) 3.0604(7) Å; N(21)–Ag(2)–N(41) 163.1(1); N(21)–Ag(2)–O(71) 106.6(1); N(41)–Ag(2)–O(71) 86.4(1)°.](/image/article/2006/DT/b605340h/b605340h-f2.gif) | ||
| Fig. 2 A view of the structure of the trifluoroacetate complex [Ag2(CF3CO2)2{C4H2S(CONMe-4-C5H4N)2}2], 8a. Selected bond parameters: Ag(1)–N(11) 2.195(2); Ag(1)–N(31) 2.182(2); Ag(1)–O(81) 2.522(2); Ag(1)–O(81A) 2.655(3); Ag(1)–Ag(1A) 3.3265(6) Å; N(31)–Ag(1)–N(11) 160.4(1); N(31)–Ag(1)–O(81) 104.9(1); N(11)–Ag(1)–O(81) 89.6(1)°. Symmetry equivalents: A, −x, 2 − y, −2 − z. For the independent molecule, corresponding parameters are: Ag(2)–N(21) 2.145(2); Ag(2)–N(41) 2.160(3); Ag(2)–O(71) 2.500(3); Ag(2)–O(71A) 2.677(3); Ag(2)–Ag(2A) 3.0604(7) Å; N(21)–Ag(2)–N(41) 163.1(1); N(21)–Ag(2)–O(71) 106.6(1); N(41)–Ag(2)–O(71) 86.4(1)°. | ||
The structure of the nitrate derivative 8b is similar to that of 8a and is shown in Fig. 3, with conformational parameters listed in Table 1. There are two independent molecules with similar structures (Fig. 3, Table 1) and they are arranged in chains with the intermolecular distance Ag(1)⋯Ag(2B) = 3.48 Å, which may represent a weak argentophilic bond.9 The nitrate ions lie above and below each macrocycle, but are only weakly bonded as κ2-µ2-nitrate [molecule 1, Ag(1)–O(71) = 2.79, Ag(1)–O(72A) = 2.77 Å; molecule 2, Ag(2)–O(63) = 2.66, Ag(2)–O(61A) = 2.68 Å] and are omitted from Fig. 3, for clarity. Short Ag⋯Ag distances with weakly bridging nitrate have been observed previously.9
![A view of the structure of the nitrate complex [Ag2(NO3)2{C4H2S(CONMe-4-C5H4N)2}2], 8b, showing the chains of macrocycles. Selected bond parameters: Ag(1)–N(1) 2.132(6); Ag(1)–N(23A) 2.150(6); Ag(1)–Ag(1A) 3.140(1) Å; N(1)–Ag(1)–N(23A) 167.6(2)°. For the independent molecule, corresponding parameters are: Ag(2A)–N(53B) 2.145(6); Ag(2A)–N(31A) 2.152(5); Ag(2A)–Ag(2B) 3.038(1) Å; N(53B)–Ag(2A)–N(31A) 166.3(2)°. Symmetry equivalents: A, −x, 1 −
y, −2 −
z; B, −x, 2 −
y, −2 −
z.](/image/article/2006/DT/b605340h/b605340h-f3.gif) | ||
| Fig. 3 A view of the structure of the nitrate complex [Ag2(NO3)2{C4H2S(CONMe-4-C5H4N)2}2], 8b, showing the chains of macrocycles. Selected bond parameters: Ag(1)–N(1) 2.132(6); Ag(1)–N(23A) 2.150(6); Ag(1)–Ag(1A) 3.140(1) Å; N(1)–Ag(1)–N(23A) 167.6(2)°. For the independent molecule, corresponding parameters are: Ag(2A)–N(53B) 2.145(6); Ag(2A)–N(31A) 2.152(5); Ag(2A)–Ag(2B) 3.038(1) Å; N(53B)–Ag(2A)–N(31A) 166.3(2)°. Symmetry equivalents: A, −x, 1 − y, −2 − z; B, −x, 2 − y, −2 − z. | ||
The structure of the triflate derivative is shown in Fig. 4, with conformational parameters in Table 1. The overall structure is similar to those of 8a (Fig. 2) and 8b (Fig. 3), but the transannular silver⋯silver distances in the two independent molecules [Ag(1)⋯Ag(1A) = 3.42 Å, Ag(2)⋯Ag(2A) = 3.60 Å] are longer than in the trifluoroacetate or nitrate derivative and can, at best, represent only very weak bonding interactions.9 The closest intermolecular distance Ag(1)⋯Ag(2) = 3.70 Å. In the molecule containing Ag(2), the κ1-µ2-triflate ions bridge weakly between the silver(I) ions [Ag(2)–O(61) 2.597(8); Ag(2)–O(61A) = 2.73(1) Å] in a similar way as the trifluoroacetate ions in 8a (Fig. 2) but, in the molecule containing Ag(1), the κ2-µ2-triflate ions bridge weakly between the silver(I) ions [Ag(1)–O(71) = 2.75(1); Ag(1)–O(72A) = 2.79(1) Å] in a similar way as the nitrate ions in 8b, indicating that there is very little difference in energy between these bridging modes.
![A view of the structure of the triflate complex [Ag2(CF3SO3)2{C4H2S(CONMe-4-C5H4N)2}2], 8c. Selected bond parameters: Ag(2)–N(31) 2.162(7); Ag(2)–N(53A) 2.172(7); Ag(2)–O(61) 2.597(8) Å; N(31)–Ag(2)–N(53A) 166.3(3); N(31)–Ag(2)–O(61) 92.3(3); N(53A)–Ag(2)–O(61) 93.3(3)°. Symmetry equivalent: A, 1 −
x, 3 −
y, 1 −
z. For the independent molecule, corresponding parameters are: Ag(1)–N(1) 2.160(6); Ag(1)–N(23A) 2.181(7) Å; N(1)–Ag(1)–N(23A) 166.5(3)°.](/image/article/2006/DT/b605340h/b605340h-f4.gif) | ||
| Fig. 4 A view of the structure of the triflate complex [Ag2(CF3SO3)2{C4H2S(CONMe-4-C5H4N)2}2], 8c. Selected bond parameters: Ag(2)–N(31) 2.162(7); Ag(2)–N(53A) 2.172(7); Ag(2)–O(61) 2.597(8) Å; N(31)–Ag(2)–N(53A) 166.3(3); N(31)–Ag(2)–O(61) 92.3(3); N(53A)–Ag(2)–O(61) 93.3(3)°. Symmetry equivalent: A, 1 − x, 3 − y, 1 − z. For the independent molecule, corresponding parameters are: Ag(1)–N(1) 2.160(6); Ag(1)–N(23A) 2.181(7) Å; N(1)–Ag(1)–N(23A) 166.5(3)°. | ||
The structure of the hexafluorophosphate derivative 8d is shown in Fig. 5 and conformational parameters are listed in Table 1. The primary structure in this case is a polymer, formed by bridging of the ligands between silver(I) atoms in a linear fashion. The hexafluorophosphate anion is not coordinated [shortest contact Ag⋯F(42A) = 2.96 Å]. Interestingly, the N-methylamide –N(Me)C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)-group of the ligand 6 exists in the unusual extended trans,trans,syn conformation (Chart 2, Table 1). In general, the N-methylamide group tends to exist in the cis conformation, that leads to a folded conformation of the molecule, while the opposite trans conformation, which leads to the more linear structure, is most commonly observed for the –NHC(
O)-group of the ligand 6 exists in the unusual extended trans,trans,syn conformation (Chart 2, Table 1). In general, the N-methylamide group tends to exist in the cis conformation, that leads to a folded conformation of the molecule, while the opposite trans conformation, which leads to the more linear structure, is most commonly observed for the –NHC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)-group.12 Although the interconversion of the cis and trans N-methylamide group in solution has been observed, this appears to be the first solid state structure of a complex containing the trans N-methylamide group.12 Two further differences between 8d and the macrocyclic complexes 8a–8c are that the carbonyl groups are roughly syn to the thiophene sulfur atom, and that the pyridyl groups are much closer to co-planarity with the amide units (Table 1). The polymer chains of 8d undergo further self-assembly in the solid state in the following way (Fig. 5). The silver atoms of one linear polymer chain are weakly coordinated to the sulfur atoms of the thiophene groups of a second polymer chain [Ag⋯S(22) = 3.32 Å], and vice-versa, to give a double stranded polymer chain. Then, further coordination of silver(I) atoms to the carbonyl atoms of a second pair of polymer chains [Ag⋯O(27) = 3.10 Å] occurs to form a supramolecular sheet structure (Fig. 5).
O)-group.12 Although the interconversion of the cis and trans N-methylamide group in solution has been observed, this appears to be the first solid state structure of a complex containing the trans N-methylamide group.12 Two further differences between 8d and the macrocyclic complexes 8a–8c are that the carbonyl groups are roughly syn to the thiophene sulfur atom, and that the pyridyl groups are much closer to co-planarity with the amide units (Table 1). The polymer chains of 8d undergo further self-assembly in the solid state in the following way (Fig. 5). The silver atoms of one linear polymer chain are weakly coordinated to the sulfur atoms of the thiophene groups of a second polymer chain [Ag⋯S(22) = 3.32 Å], and vice-versa, to give a double stranded polymer chain. Then, further coordination of silver(I) atoms to the carbonyl atoms of a second pair of polymer chains [Ag⋯O(27) = 3.10 Å] occurs to form a supramolecular sheet structure (Fig. 5).
![Views of the structure of the hexafluorophosphate derivative [Ag{C4H2S(CONMe-4-C5H4N)2}]n[PF6]n, 8d: above, the cationic building block unit; below, part of the supramolecular sheet structure. Selected bond parameters: Ag–N(11) 2.131(4); Ag–N(33A) 2.140(4) Å; N(11)–Ag–N(33A) 168.6(2)°. Symmetry equivalents: A, 0.5 −
x, −0.5 −
y, 1.5 −
z; D, −x, 1 −
y, 1 −
z.](/image/article/2006/DT/b605340h/b605340h-f5.gif) | ||
| Fig. 5 Views of the structure of the hexafluorophosphate derivative [Ag{C4H2S(CONMe-4-C5H4N)2}]n[PF6]n, 8d: above, the cationic building block unit; below, part of the supramolecular sheet structure. Selected bond parameters: Ag–N(11) 2.131(4); Ag–N(33A) 2.140(4) Å; N(11)–Ag–N(33A) 168.6(2)°. Symmetry equivalents: A, 0.5 − x, −0.5 − y, 1.5 − z; D, −x, 1 − y, 1 − z. | ||
From the above, it is clear that the solid state structures are anion dependent, and it was of interest to know if this is also true for the structures in solution.6 The complexes are sparingly soluble in non-polar solvents, but complexes 8a–8c are sufficiently soluble in methanol to give satisfactory 1H NMR spectra and ESI mass spectra. The 1H NMR spectra, obtained in solution in CD3OD, are similar to the spectrum of the free ligand 6, with only modest coordination shifts observed. There was no evidence for the presence of isomeric mixtures, but the data are inconclusive because rapid exchange might give coalescence of signals. Satisfactory low temperature NMR spectra could not be obtained because the solubility was too low.
The ESI-MS of the complexes 8a–8c, obtained in dilute solution in methanol, showed the highest mass peak envelope centered at m/z = 1033, 982, and 1071, corresponding to the ions [62 + 107Ag2 + X]+, with X = CF3CO2, NO3 and CF3SO3, respectively, consistent with the presence of macrocycles in solution. The ESI-MS of complex 8d, obtained using a dilute solution in acetonitrile at low temperature, contained an envelope of peaks at m/z = 1065 corresponding to [62 + 107Ag2 + X]+, where X = PF6, but also contained higher mass peaks centred at m/z = 1671 and 2276 corresponding to [63 + 107Ag3 + X2]+ and [64 + 107Ag4 + X3]+, indicating the presence of at least some trimer and tetramer along with the macrocyclic dimer in solution. The ESI-MS of 8a–8c obtained under similar conditions failed to show peaks in these regions, the highest mass peaks being assigned to dimers as described above.
The ligand SC4H2-2,5-{C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/h3_char_e001.gif) O)NHCH2-3-C5H4N}2, 7
O)NHCH2-3-C5H4N}2, 7
The ligand SC4H2-2,5-{C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)NHCH2-3-C5H4N}2, 7, was prepared by reaction of 3-(aminomethyl)pyridine with the acid chloride derivative 2,5-C4H2S[C(
O)NHCH2-3-C5H4N}2, 7, was prepared by reaction of 3-(aminomethyl)pyridine with the acid chloride derivative 2,5-C4H2S[C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)OCl]2, and its structure is shown in Fig. 6 with conformational parameters listed in Table 1. The amide units adopt the usual trans C(O)NH conformation and the carbonyl groups are roughly syn to the thiophene sulfur atom. Each ligand is involved in four intermolecular hydrogen bonds, of which two are N(pyridyl)⋯HN bonds and two are C
O)OCl]2, and its structure is shown in Fig. 6 with conformational parameters listed in Table 1. The amide units adopt the usual trans C(O)NH conformation and the carbonyl groups are roughly syn to the thiophene sulfur atom. Each ligand is involved in four intermolecular hydrogen bonds, of which two are N(pyridyl)⋯HN bonds and two are C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O⋯HN bonds, to form a sheet structure (Fig. 6).
O⋯HN bonds, to form a sheet structure (Fig. 6). | ||
| Fig. 6 The sheet structure of the ligand C4H2S(CONHCH2-3-C5H4N)2, 7. Hydrogen bonding distances N(18)⋯N(32A) 3.022(6); N(28)⋯O(20B) 2.953(5) Å. Symmetry equivalents: A, x, y + 1, z + 1; B, x − 1, y − 1, z; C, x + 1, y + 1, z; E, x, y − 1, z − 1. | ||
The silver(I) complex 9
The only silver(I) complex of ligand 7 to form good single crystals was the polymeric derivative [Ag(µ-7)]n[CF3CO2]n, 9, as shown in Scheme 2. | ||
| Scheme 2 | ||
The structure of complex 9 is shown in Fig. 7, with conformational parameters in Table 1.
 | ||
Fig. 7 The structure of the polymeric complex [Ag(CF3CO2){C4H2S(CONHCH2-3-C5H4N)2}n, 9: above, part of the double stranded polymer formed by Ag⋯Ag interactions (trifluoroacetate groups are omitted for clarity); below, the sheet structure formed through further supramolecular association involving C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O⋯HN and Ag–O(trifluoroacetate)⋯HN hydrogen bonds (fluorine atoms of the trifluoroacetate groups are omitted for clarity). Selected bond parameters: Ag–N(11) 2.170(8); Ag–N(32B) 2.208(8); Ag⋯O(41C) 2.63(1); Ag–AgA 3.176(2) Å; N(11)–Ag–N(32B) 156.9(3)°; hydrogen bonding distances: N(18)⋯O(27D) 2.86(1); N(28)⋯O(41) 2.77(1) Å. Symmetry equivalents: A, 3 −
x, 2 −
y, 2 −
z; B, x + 2, y, z + 1. O⋯HN and Ag–O(trifluoroacetate)⋯HN hydrogen bonds (fluorine atoms of the trifluoroacetate groups are omitted for clarity). Selected bond parameters: Ag–N(11) 2.170(8); Ag–N(32B) 2.208(8); Ag⋯O(41C) 2.63(1); Ag–AgA 3.176(2) Å; N(11)–Ag–N(32B) 156.9(3)°; hydrogen bonding distances: N(18)⋯O(27D) 2.86(1); N(28)⋯O(41) 2.77(1) Å. Symmetry equivalents: A, 3 −
x, 2 −
y, 2 −
z; B, x + 2, y, z + 1. | ||
In complex 9, the conformation of the ligand is similar to that of the free ligand 7, except for differences in rotation of the pyridylmethyl groups (Table 1, Fig. 6 and 7). This conformation is naturally suited to form the observed polymeric complex 9, with the intrachain separation Ag⋯Ag = 18.2 Å. The polymers undergo supramolecular association to form a sheet structure, by a combination of secondary bonding interactions. First, pairs of polymer chains associate through Ag⋯Ag interactions to form a double stranded polymer [Ag⋯Ag = 3.176(2) Å, Fig. 7, top]. Next, the double stranded polymers associate through forming weak coordinate bonds and hydrogen bonds to neighboring chains. Thus, each ligand forms two C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O⋯HN hydrogen bonds (one as donor and one as acceptor), and also an Ag⋯O⋯HN hydrogen bond involving a trifluoroacetate anion, which, through a single oxygen atom O(41), coordinates to a silver atom in one chain [Ag⋯O(41) 2.63(1) Å] and forms a hydrogen bond to an NH group of a ligand in a neighboring chain [N(28)⋯O(41) = 2.77(1) Å, Fig. 7, bottom). There are no secondary bonding interactions of the type (thiophene)S⋯Ag.
O⋯HN hydrogen bonds (one as donor and one as acceptor), and also an Ag⋯O⋯HN hydrogen bond involving a trifluoroacetate anion, which, through a single oxygen atom O(41), coordinates to a silver atom in one chain [Ag⋯O(41) 2.63(1) Å] and forms a hydrogen bond to an NH group of a ligand in a neighboring chain [N(28)⋯O(41) = 2.77(1) Å, Fig. 7, bottom). There are no secondary bonding interactions of the type (thiophene)S⋯Ag.
Discussion
For the silver(I) complexes 8 there is an easy equilibrium in solution between macrocyclic and ring-opened complexes, as shown in Scheme 1. The equilibrium is made possible through flexibility of the ligands, especially by rotation of the thiophene and amide units (Table 1). The macrocyclic complexes crystallize for the trifluoroacetate, nitrate and triflate derivatives 8a–8c, whereas the polymer crystallizes for the hexafluorophosphate derivative 8d. The macrocyclic isomer appears to be stabilized by weak bridging of the oxygen donor anion between the two silver(I) centers in 8a–8c, and these oxygen-donor bridges also enhance a transannular secondary Ag⋯Ag bonding interaction in complexes 8a [Ag⋯Ag = 3.3266(6), 3.0604(7) Å] and 8b [Ag⋯Ag = 3.140(1), 3.038(1) Å], which contain the relatively stronger trifluoroacetate or nitrate donors. The complex 8c, with the weaker triflate donors, has a longer transannular distance [Ag⋯Ag = 3.42, 3.60 Å]. In complexes 8, the macrocycles pack in chains (see Fig. 3) and there are somewhat short contacts between silver(I) ions of neighboring macrocycles [Ag⋯Ag = 3.55, 3.48, 3.70 Å in 8a, 8b, 8c respectively], but even the shortest of these can represent only a weak bonding interaction. The complexes 8a and 8b are isomorphous and the chains of silver(I) ions lie parallel to the b-axis; the shorter Ag⋯Ag contacts for the nitrate derivative 8b lead to a significantly shorter cell parameter [b = 12.935(2) and 14.0366(5) Å in 8b and 8a, respectively]. In the polymeric complex 8d, the interchain association in the solid state occurs through weak Ag⋯S and Ag⋯O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bonding interactions and not through secondary Ag⋯Ag bonding interactions (Fig. 5). A summary of the Ag⋯Ag distances is given in Table 2.
C bonding interactions and not through secondary Ag⋯Ag bonding interactions (Fig. 5). A summary of the Ag⋯Ag distances is given in Table 2.
| Complex | Intramolecular Ag⋯Ag | Intermolecular Ag⋯Ag |
|---|---|---|
| 8a | 3.33 | 3.55 |
| 3.06 | ||
| 8b | 3.14 | 3.48 |
| 3.04 | ||
| 8c | 3.42 | 3.70 |
| 3.60 | ||
| 9 | 3.18 |
The preferred trans stereochemistry of the amide unit in ligand 7 (Fig. 6) naturally gives a greater tendency to polymer formation with silver(I), when compared to ligand 6. Only one complex crystallized well, but the trifluoroacetate complex 9 was indeed a polymer and not a macrocycle. Complex 9 is of interest because of the many potential forms of secondary bonding that could be formed. The structure (Fig. 7) reveals the presence of Ag⋯Ag argentophilic bonds, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O⋯HN and CF3C(
O⋯HN and CF3C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)O⋯HN hydrogen bonds, and CF3C(
O)O⋯HN hydrogen bonds, and CF3C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)O⋯Ag secondary coordinate bonds, but no secondary bonding interactions of the type (thiophene)S⋯Ag. From the structural data alone, it is not clear what the relative strengths of the individual secondary bonding interactions are, and it is presumed that the polymer chains are arranged so as to give the maximum combined secondary bonding interactions. The outcome is different in the packing of the polymer chains of 8d [Ag⋯S, but no Ag⋯Ag] and 9 [Ag⋯Ag, but no Ag⋯S].
O)O⋯Ag secondary coordinate bonds, but no secondary bonding interactions of the type (thiophene)S⋯Ag. From the structural data alone, it is not clear what the relative strengths of the individual secondary bonding interactions are, and it is presumed that the polymer chains are arranged so as to give the maximum combined secondary bonding interactions. The outcome is different in the packing of the polymer chains of 8d [Ag⋯S, but no Ag⋯Ag] and 9 [Ag⋯Ag, but no Ag⋯S].
Experimental
1H and 13C{1H} spectra were recorded using a Varian Mercury 400 spectrometer; chemical shifts are reported relative to TMS. The proton labeling system for assignment of the NMR spectra is described in Chart 3. ESI mass spectra were recorded using a Micromass LCT spectrometer and were calibrated with NaI at a concentration of 2 µg µL−1 in 50 : 50 propan-2-ol : water. CSI mass spectra were recorded using the same spectrometer but at a desolvation temperature of 0 to −30 °C and were calibrated with CsI at a concentration of 10 µg µL−1 in MeCN solution. Natural abundance of silver isotopes is 107Ag 51.8%; 109Ag 48.2%. Silver complexes were handled in subdued light to avoid photolysis.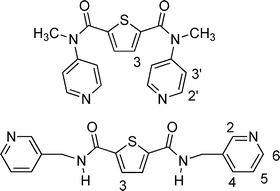 | ||
| Chart 3 | ||
2,5-C4H2S[C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/h3_char_e001.gif) O)N(Me)-4-C5H4N]2, 6
O)N(Me)-4-C5H4N]2, 6
To a solution of 4-(methylamino)pyridine (2.0 g, 18.5 mmol) and triethylamine (3.86 mL; 27.7 mmol) in dry CH2Cl2 (100 mL) was added a solution of 2,5-C4H2S[C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)Cl]2 (1.93 g; 9.25 mmol) in dry CH2Cl2 (20 mL). The reaction mixture was stirred at room temperature under a nitrogen atmosphere overnight, and the solvent was evaporated under vacuum to give a white solid, which was crystallized from benzene and dried under vacuum. Yield: 1.05 g (32%). NMR in CD2Cl2/CD3OD: δ(1H) = 8.49 [dd, 4H, JHH = 2 Hz, 5 Hz, H2’]; 7.21 [dd, 4H, JHH = 2 Hz, 5 Hz, H3’]; 6.68 [s, 2H, H3]; 3.43 [s, 6H, CH3]. δ(13C) = 163.5 (C
O)Cl]2 (1.93 g; 9.25 mmol) in dry CH2Cl2 (20 mL). The reaction mixture was stirred at room temperature under a nitrogen atmosphere overnight, and the solvent was evaporated under vacuum to give a white solid, which was crystallized from benzene and dried under vacuum. Yield: 1.05 g (32%). NMR in CD2Cl2/CD3OD: δ(1H) = 8.49 [dd, 4H, JHH = 2 Hz, 5 Hz, H2’]; 7.21 [dd, 4H, JHH = 2 Hz, 5 Hz, H3’]; 6.68 [s, 2H, H3]; 3.43 [s, 6H, CH3]. δ(13C) = 163.5 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 152.8, 151.4, 141.7, 132.0, 122.6, 38.5 (CH3). Anal. Calcd for C18H16N4O2S: C, 61.35; H, 4.58; N, 15.90. Found: C, 61.80; H, 4.55; N, 15.90%. Colourless crystals of 6·2HCl·4H2O were grown from a concentrated solution in CH2Cl2/Me2CO/MeOH/H2O.
O), 152.8, 151.4, 141.7, 132.0, 122.6, 38.5 (CH3). Anal. Calcd for C18H16N4O2S: C, 61.35; H, 4.58; N, 15.90. Found: C, 61.80; H, 4.55; N, 15.90%. Colourless crystals of 6·2HCl·4H2O were grown from a concentrated solution in CH2Cl2/Me2CO/MeOH/H2O.[Ag2(µ-6)2][CF3CO2]2, 8a
To a solution of [AgO2CCF3] (0.018 g; 0.082 mmol) in EtOAc (5 mL) was added a solution of the ligand 6 (0.029 g; 0.082 mmol) in CH2Cl2 (5 mL). The solution was stirred overnight and excess hexane (10 mL) was added to precipitate the product as a white solid, which was collected by filtration, washed with pentane and dried under vacuum. Yield: 0.042 g (45%). NMR in CD3OD: δ(1H) = 8.57 [d, 8H, JHH = 6 Hz, H2’], 7.53 [dd, 8H, JHH = 2 Hz, 6 Hz, H3’], 7.05 [s, 4H, H3], 3.52 [s, 12H, CH3]. Anal. Calcd for C40H32Ag2F6N8O8S2.2CH2Cl2: C: 38.32; H: 2.76; N: 8.51%. Found: C: 38.41; H: 2.33; N: 9.19%. Colourless prism crystals of 8a·CH2Cl2 were grown by slow diffusion of a solution of 6 in CH2Cl2 into a solution of AgO2CCF3 in acetone.Similarly were prepared: [Ag2(µ-6)2][NO3]2, 8b, using [AgNO3] (0.014 g; 0.085 mmol). Yield: 0.036 g (41%). NMR in CD3OD: δ(1H) = 8.62 [d, 8H, JHH = 7 Hz, H2’], 7.82 [d, 8H, JH–H = 7 Hz, H3’], 7.31 [s, 4H, H3], 3.64 [s, 12H, CH3]. Anal. Calcd for C36H32Ag2N10O10S2·3CH2Cl2: C, 36.05; H, 2.95; N, 10.78%. Found: C, 35.64; H, 2.69; N, 11.67%. Colourless plates of 8b·CH2Cl2 were grown by slow diffusion of a concentrated solution of 6 in CH2Cl2 into a solution of AgNO3 in MeOH/thf. [Ag2(µ-6)2][CF3SO3]2, 8c from [AgO3SCF3] (0.022 g; 0.085 mmol). Yield: 0.042 g (40%). NMR in CD3OD: δ(1H) = 8.57 [d, 8H, H2’, JHH = 5 Hz], 7.63 [dd, 8H, JHH = 2 Hz, 5 Hz, H3’], 7.10 [s, 4H, H3], 3.56 [s, 12H, CH3]. Anal. Calcd for C38H32Ag2F6N8O10S4: C: 37.45; H: 2.65; N: 9.19%. Found: C: 36.94; H: 2.47; N: 8.87%. Colourless block crystals of 8c·acetone were grown by slow diffusion from a concentrated solution of 6 in CH2Cl2 into a solution of [AgO3SCF3] in acetone.
[Ag(µ-6)]n[PF6]n, 8d
To a solution of AgPF6 (0.030 g, 0.119 mmol) in EtOAc (5 mL) was added a solution of 6 (0.421 g, 0.119 mmol) in CH2Cl2 (5 mL). The solution was stirred overnight to give a white solid precipitate, which was collected by filtration, washed with pentane and dried under vacuum. Yield: 0.053 g (73%). NMR in dmf-d7: δ(1H) = 8.70 [m, 4H, JHH = 2 Hz, 5 Hz, H2′]; 7.50 [m, 4H, JHH = 2 Hz, 5 Hz, H3′]; 6.93 [s, 2H, H3]; 3.48 [s, 6H, CH3]. Anal. Calcd for C18H16AgF6N4O2PS: C, 35.30; H, 2.66; N, 9.26%. Found: C, 35.72; H, 2.21; N, 8.88%. Colourless petals of 8d·2CH2Cl2 were grown by slow diffusion of a concentrated solution of AgPF6 in CH2Cl2 into a solution containing an equimolar amount of ligand 6 in methanol.2,5-C4H2S[C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/h3_char_e001.gif) O)NH–CH2-3-C5H4N]2, 7
O)NH–CH2-3-C5H4N]2, 7
To a solution mixture of 3-(aminomethyl)pyridine (2.1 g; 19.1 mmol) and excess of triethylamine (4.0 mL; 28.7 mmol) in dried THF (100 mL), was added a solution of 2,5-C4H2S[C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)OCl]2 (2.0 g; 9.56 mmol) in dried THF (20 mL). The reaction mixture was stirred at room temperature under nitrogen overnight. The solvent was evaporated under vacuum. The white solid residue was washed with water, then filtered and dried under vacuum overnight. Yield: 2.94 g (92%). NMR in dmf-d7: δ(1H) = 9.26 [t, 2H, JHH = 6 Hz, CONH]; 8.63 [d, 2H, JHH = 2 Hz, H2]; 8.50 [dd, 2H, JHH = 2 Hz, 5 Hz, H6]; 7.87 [s, 2H, H3], 7.80 [m, 2H, JHH = 2 Hz, 8 Hz, H4], 7.38 [dd, 2H, JHH = 5 Hz, 8 Hz, H5], 4.59 [d, 4H, JHH = 6 Hz, CH2]. δ(13C) = 161.8 (C
O)OCl]2 (2.0 g; 9.56 mmol) in dried THF (20 mL). The reaction mixture was stirred at room temperature under nitrogen overnight. The solvent was evaporated under vacuum. The white solid residue was washed with water, then filtered and dried under vacuum overnight. Yield: 2.94 g (92%). NMR in dmf-d7: δ(1H) = 9.26 [t, 2H, JHH = 6 Hz, CONH]; 8.63 [d, 2H, JHH = 2 Hz, H2]; 8.50 [dd, 2H, JHH = 2 Hz, 5 Hz, H6]; 7.87 [s, 2H, H3], 7.80 [m, 2H, JHH = 2 Hz, 8 Hz, H4], 7.38 [dd, 2H, JHH = 5 Hz, 8 Hz, H5], 4.59 [d, 4H, JHH = 6 Hz, CH2]. δ(13C) = 161.8 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 150.0, 149.1, 144.1, 136.0, 135.6, 129.0, 124.1, 41.4 (CH2). Anal. Calcd for C18H16N4O2S: C, 61.35; H, 4.58; N, 15.90. Found: C, 60.94; H, 4.39; N, 15.93%. Colourless crystals of [(NC5H4)CH2N(H)C(O)(C4H2S)C(O)N(H)CH2(C5H4N)] were grown by slow diffusion from a concentrated solution of the ligand in a mixture of dichloromethane and methanol to THF.
O), 150.0, 149.1, 144.1, 136.0, 135.6, 129.0, 124.1, 41.4 (CH2). Anal. Calcd for C18H16N4O2S: C, 61.35; H, 4.58; N, 15.90. Found: C, 60.94; H, 4.39; N, 15.93%. Colourless crystals of [(NC5H4)CH2N(H)C(O)(C4H2S)C(O)N(H)CH2(C5H4N)] were grown by slow diffusion from a concentrated solution of the ligand in a mixture of dichloromethane and methanol to THF.[Ag2(µ-2,5-C4H2S[C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/h3_char_e001.gif) O)NH–CH2-3-C5H4N]2)2](CF3CO2)2, 9
O)NH–CH2-3-C5H4N]2)2](CF3CO2)2, 9
To a solution of [AgO2CCF3] (0.03 g; 0.136 mmol) in dichloromethane (5 mL), was added a solution of 2,5-C4H2S[C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)NH–CH2-3-C5H4N]2 (0.048 g; 0.136 mmol) in dmf (2 mL), to give a white precipitate. The solution mixture was stirred overnight and excess of pentane (10 mL) was added to precipitate the product. A white solid was collected by filtration, washed with acetone and dried under vacuum. Yield: 0.073 g (46%). NMR in dmf-d7: δ(1H) = 9.36 [t, 2H, JHH = 6 Hz, CONH]; 8.71 [d, 2H, JHH = 2 Hz, H2]; 8.60 [d, 2H, JHH = 5 Hz, H6]; 7.95 [m, 2H, JHH = 2 Hz, 5 Hz, 8 Hz, H4]; 7.87 [s, 2H, H3]; 7.52 [dd, 2H, JHH = 5 Hz, 8 Hz, H5]; 4.62 [d, 4H, JHH = 6 Hz, CH2]. Anal. Calcd for C41H35Ag2F6N8O8S2: C, 42.39; H, 3.04; N, 9.65. Found: C, 42.54; H, 2.80; N, 10.03%. Colourless plates of [Ag{µ(NC5H4)CH2N(H)C(O)(SC4H2)C(O)N(H)CH2(C5H4N)}] [CF3CO2]·0.5 acetone were grown by slow diffusion from a concentrated solution of the ligand 7 in a mixture of dichloromethane and methanol to a concentrated solution of silver trifluoroacetate in acetone.
O)NH–CH2-3-C5H4N]2 (0.048 g; 0.136 mmol) in dmf (2 mL), to give a white precipitate. The solution mixture was stirred overnight and excess of pentane (10 mL) was added to precipitate the product. A white solid was collected by filtration, washed with acetone and dried under vacuum. Yield: 0.073 g (46%). NMR in dmf-d7: δ(1H) = 9.36 [t, 2H, JHH = 6 Hz, CONH]; 8.71 [d, 2H, JHH = 2 Hz, H2]; 8.60 [d, 2H, JHH = 5 Hz, H6]; 7.95 [m, 2H, JHH = 2 Hz, 5 Hz, 8 Hz, H4]; 7.87 [s, 2H, H3]; 7.52 [dd, 2H, JHH = 5 Hz, 8 Hz, H5]; 4.62 [d, 4H, JHH = 6 Hz, CH2]. Anal. Calcd for C41H35Ag2F6N8O8S2: C, 42.39; H, 3.04; N, 9.65. Found: C, 42.54; H, 2.80; N, 10.03%. Colourless plates of [Ag{µ(NC5H4)CH2N(H)C(O)(SC4H2)C(O)N(H)CH2(C5H4N)}] [CF3CO2]·0.5 acetone were grown by slow diffusion from a concentrated solution of the ligand 7 in a mixture of dichloromethane and methanol to a concentrated solution of silver trifluoroacetate in acetone.X-Ray data collection and reduction
Crystals were mounted on glass fibers, and data were collected using a Nonius Kappa-CCD diffractometer with COLLECT (Nonius, 1998) software. Crystal cell refinement and data reduction were carried out using the Nonius DENZO package. The data were scaled using SCALEPACK (Nonius, 1998). The SHELXTL-NT V6.1 (Sheldrick, G. M., Madison, WI) program package was used to solve and refine the structures by direct methods. The hydrogen atom positions were calculated geometrically and were included as riding on their respective carbon atoms. Details of the data collection and refinement are given in Tables 3 and 4. The figures show 20% probability ellipsoids.6·2HCl·4H2O: There is a plane of symmetry passing through the centre of the molecule and containing the oxygen atoms of the water molecules.
| Compound | 6·2HCl·4H2O | 7 | 9.0.5acetone |
|---|---|---|---|
| Formula | C18H26Cl2N4O6S | C18H16N4O2S | C21.5H19AgF3N4O4.5S |
| FW | 497.39 | 352.41 | 602.34 |
| Temp./K | 150(2) | 150(2) | 150(2) |
| λ/Å | 0.71073 | 0.71073 | 0.71073 |
| Cryst. syst. | Orthorhombic | Triclinic | Triclinic |
| Space gp. cell dimens. | Pnma | P-1 | P-1 |
| a/Å | 8.3269(2) | 6.077(1) | 7.706(1) |
| b/Å | 14.7324(5) | 7.346(1) | 12.748(2) |
| c/Å | 19.1904(5) | 9.783(2) | 14.185(2) |
| α/° | 90 | 102.253(8) | 112.983(7) |
| β/° | 90 | 91.961(9) | 104.112(6) |
| γ/° | 90 | 109.728(8) | 94.866(8) |
| V/Å3 | 2354.2(1) | 399.1(1) | 1218.6(3) |
| Z | 4 | 1 | 2 |
| d(calc)/Mg m−3 | 1.403 | 1.466 | 1.642 |
| Abs. Coeff./mm−1 | 0.405 | 0.224 | 0.973 |
| F(000) | 1040 | 184 | 604 |
| Ind. Refl. | 2162 | 1870 | 3492 |
| Abs. Corr. | Integration | Integration | Integration |
| Data/restr./param. | 2162/10/164 | 1870/3/227 | 3492/0/335 |
| Goof | 1.109 | 1.136 | 1.078 |
| R1 [I > 2σ(I)] | 0.087 | 0.043 | 0.064 |
| wR2 [I > 2σ(I)] | 0.259 | 0.106 | 0.136 |
| Compound | 8a·CH2Cl2 | 8b·CH2Cl2 | 8c·acetone | 8d·2CH2Cl2 |
|---|---|---|---|---|
| Formula | C41H34Ag2Cl2F6N8O8S2 | C37H34Ag2Cl2N10O10S2 | C41H38Ag2F6N8O11S4 | C20H20AgCl4F6N4O2PS |
| FW | 1231.52 | 1129.50 | 1276.77 | 775.10 |
| Temp./K | 150(2) | 150(2) | 150(2) | 150(2) |
| λ/Å | 0.71073 | 0.71073 | 0.71073 | 0.71073 |
| Cryst. syst. | Triclinic | Triclinic | Triclinic | Monoclinic |
| Space gp. cell dimens. | P-1 | P-1 | P-1 | P21/n |
| a/Å | 11.6130(5) | 11.644(1) | 13.4354(5) | 11.5598(3) |
| b/Å | 14.0366(5) | 12.935(2) | 13.9401(7) | 19.7777(5) |
| c/Å | 16.2974(6) | 15.592(2) | 16.2085(9) | 12.8335(3) |
| α/° | 65.287(2) | 68.512(6) | 108.136(2) | 90 |
| β/° | 81.037(2) | 76.221(7) | 97.541(3) | 109.256(2) |
| γ/° | 72.264(2) | 73.795(6) | 117.121(2) | 90 |
| V/Å3 | 2297.3(1) | 2074.0(4) | 2430.1(2) | 2769.9(1) |
| Z | 2 | 2 | 2 | 4 |
| d(calc)/Mg m−3 | 1.780 | 1.809 | 1.745 | 1.859 |
| Abs. Coeff./mm−1 | 1.146 | 1.245 | 1.067 | 1.315 |
| F(000) | 1228 | 1132 | 1280 | 1536 |
| Ind. Refl. | 10374 | 7138 | 8518 | 4883 |
| Abs. Corr. | Integration | Integration | Integration | Integration |
| Data/restr./param. | 10374/0/623 | 7138/0/568 | 8518/0/649 | 4883/0/352 |
| Goof | 1.060 | 0.958 | 1.037 | 1.073 |
| wR1 [I > 2σ(I)] | 0.041 | 0.062 | 0.076 | 0.049 |
| wR2 [I > 2σ(I)] | 0.090 | 0.150 | 0.205 | 0.134 |
7: The molecule is chiral in space group P-1 and the correct choice was confirmed by the absolute structure parameter.
8a·CH2Cl2, 8b·CH2Cl2 and 8c·acetone: There are two independent macrocycles, each of which contain a centre of symmetry.
8d·2CH2Cl2: No unusual features were observed.
9·0.5acetone: The crystal was twinned and the twinning was treated using the program ROTAX.
CCDC reference numbers 604509–604515.
For crystallographic data in CIF or other electronic format see DOI: 10.1039/b605340h
Acknowledgements
We thank the NSERC (Canada) for financial support.References
- Review: S. L. James, Macromol. Symp., 2004, 209, 119 Search PubMed.
- (a) S.-Y. Su, A. M. Goforth, M. D. Smith and H.-C. zur Loye, Inorg. Chem., 2003, 42, 5685 CrossRef CAS; (b) D. M. Shin, I. S. Lee, Y.-A. Lee and Y. K. Chung, Inorg. Chem., 2003, 42, 2977 CrossRef CAS; (c) P. Miller, M. Nieuwenhuyzen, J. P. H. Charmant and S. L. James, CrystEngComm, 2004, 6, 408 RSC; (d) T. J. Burchell and R. J. Puddephatt, Inorg. Chem., 2005, 44, 3718 CrossRef CAS; (e) J. M. J. Paulusse and R. P. Sijbesma, Chem. Commun., 2003, 1494 RSC.
- (a) H.-L. Hu, C.-Y. Yeh and J.-D. Chen, Eur. J. Inorg. Chem., 2004, 4696 CrossRef CAS; (b) I. S. Lee, D. M. Shin and Y. K. Chung, Chem.–Eur. J., 2004, 10, 3158 CrossRef CAS; (c) R. P. Feazell, C. E. Carson and K. K. Klausmeyer, Inorg. Chem., 2005, 44, 996 CrossRef CAS; (d) L. Carlucci, G. Ciani, D. M. Proserpio and L. Spadacini, CrystEngComm, 2004, 6, 96 RSC; (e) X.-C. Huang, J.-P. Zhang, Y.-Y. Lin and X.-M. Chen, Chem. Commun., 2005, 2232 RSC; (f) D. Braga, M. Curzi, F. Grepioni and M. Polito, Chem. Commun., 2005, 2915 RSC.
- (a) N. Masciocchi, G. A. Ardizzoia, G. LaMonica, A. Maspero and A. Sironi, Angew. Chem., Int. Ed., 1998, 37, 3366 CrossRef CAS; (b) L. Carlucci, G. Ciani, D. M. Proserpio and A. Sironi, Inorg. Chem., 1998, 37, 5941 CrossRef CAS.
- N. L. S. Yue, M. C. Jennings and R. J. Puddephatt, Chem. Commun., 2005, 4792 RSC.
- (a) C. L. Chen, B. S. Kang and C. Y. Su, Aust. J. Chem., 2006, 59, 3 CrossRef CAS; (b) H.-P. Wu, C. Janiak, G. Rheinwald and H. Lang, J. Chem. Soc., Dalton Trans., 1999, 183 RSC; (c) M. A. Withersby, A. J. Blake, N. R. Champness, P. Hubberstey, W.-S. Li and M. Schroder, Angew. Chem., Int. Ed. Engl., 1997, 36, 2327 CrossRef CAS; (d) C. Janiak, L. Uehlin, H.-P. Wu, P. Klufers, H. Piotrowski and T. G. Scharmann, J. Chem. Soc., Dalton Trans., 1999, 3121 RSC.
- N. L. S. Yue, M. C. Jennings and R. J. Puddephatt, Inorg. Chem., 2005, 44, 1125 CrossRef CAS.
- (a) Z. Qin, M. C. Jennings and R. J. Puddephatt, Chem. Commun., 2002, 354–355 RSC; (b) T. J. Burchell, D. J. Eisler, M. C. Jennings and R. J. Puddephatt, Chem. Commun., 2003, 2228–2229 RSC; (c) T. J. Burchell, D. J. Eisler and R. J. Puddephatt, Chem. Commun., 2004, 944–945 RSC.
- (a) M. A. Omary, T. R. Webb, Z. Assefa, G. E. Shankle and H. H. Patterson, Inorg. Chem., 1998, 37, 1380 CrossRef CAS; (b) M. B. Duriska, S. R. Batten and D. J. Price, Aust. J. Chem., 2006, 59, 26 CrossRef CAS; (c) M. Du, C.-P. Li and X.-J. Zhao, Cryst. Growth Des., 2006, 6, 335 CrossRef CAS; (d) T. Dorn, K. M. Fromm and C. Janiak, Aust. J. Chem., 2006, 59, 22 CrossRef CAS; (e) O.-S. Jung, Y. J. Kim, Y.-A. Lee, S. W. Kang and S. N. Choi, Cryst. Growth Des., 2004, 4, 23 CrossRef CAS.
- (a) Y.-B. Dong, Y. Geng, J.-P. Ma and R.-Q. Huang, Organometallics, 2006, 25, 447 CrossRef CAS; (b) L. R. Hanton, C. Richardson, W. T. Robinson and J. M. Turnbull, Chem. Commun., 2000, 2465 RSC; (c) M. Ghassemzadeh, A. Sharifi, J. Malakootikhah, B. Neumuller and E. Iravani, Inorg. Chim. Acta, 2004, 357, 2445; (d) E. Virtanen, J. Koivukorpi, J. Tamminen, P. Manttari and E. Kolehmainen, J. Organomet. Chem., 2003, 668, 43 CrossRef CAS.
- (a) H. Goto, Y. Okamoto and E. Yashima, Chem.–Eur. J., 2002, 8, 4027 CrossRef CAS; (b) L. M. Goldenberg, P. J. Skabara, D. M. Roberts, R. Berridge, E. Orti, P. M. Viruela and R. Pou-Amerigo, J. Mater. Chem., 2000, 10, 2458 RSC; (c) J. Bukowska and K. Jackowska, Synth. Met., 2000, 35, 135; (d) E. A. Bazzaoui, S. Aeiyach, J. Aubard, N. Felidj, G. Lévi, N. Sakmeche and P. C. Lacaze, J. Chim. Phys. Phys.-Chim. Biol., 1998, 95, 1526 CrossRef CAS.
- (a) F. D. Lewis, T. M. Long, C. L. Stern and W. Liu, J. Phys. Chem. A, 2003, 107, 3254 CrossRef CAS; (b) M. Jørgensen and F. C. Krebs, Tetrahedron Lett., 2001, 42, 4717 CrossRef CAS; (c) I. Azumaya, I. Okamoto, S. Nakayama, A. Tanatani, K. Yamaguchi, K. Shudo and H. Kagechika, Tetrahedron, 1999, 55, 11237 CrossRef CAS; (d) F. C. Krebs, M. Larsen, M. Jørgensen, P. R. Jensen, M. Bielecki and K. Schaumburg, J. Org. Chem., 1998, 63, 9872 CrossRef CAS; (e) I. Azumaya, K. Yamaguchi, I. Okamoto, H. Kagechika and K. Shudo, J. Am. Chem. Soc., 1995, 117, 9083 CrossRef CAS; (f) I. Azumaya, H. Kagechika, K. Yamaguchi and K. Shudo, Tetrahedron, 1995, 51, 5277 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2006 |
