“Click” synthesis of small molecule probes for activity-based fingerprinting of matrix metalloproteases†
Jun
Wang
a,
Mahesh
Uttamchandani
b,
Junqi
Li
a,
Mingyu
Hu
a and
Shao Q.
Yao
*abc
aDepartment of Chemistry, National University of Singapore, 3 Science Drive 3, Singapore 117543. E-mail: chmyaosq@nus.edu.sg; Fax: (+65) 67791691; Tel: (+65) 68741683
bDepartment of Biological Sciences, National University of Singapore, Singapore 117543
cNUS MedChem Program of the Office of Life Sciences, National University of Singapore, Singapore 117543
First published on 15th August 2006
Abstract
By using “Click Chemistry”, we achieved the facile synthesis of various affinity-based hydroxamate probes that enable generation of activity-based fingerprints of a variety of metalloproteases, including matrix metalloproteases (MMPs), in proteomics experiments.
Enzymes play a key role in virtually every biological process. They have long been considered valuable drug targets for potential treatments of major human diseases. Matrix metalloproteases (MMPs), for example, are a family of zinc-containing proteases which represent a family of at least 23 members in humans alone, and have become attractive targets for drug discovery because of their implication in diseases such as arthritis, Alzheimer's disease, cancer, and heart disease.1 Despite their well-documented pro-tumorigenic actions, only three MMPs, namely MMP-1, -2 and -7, have thus far been experimentally validated as potential cancer targets. Another three (i.e. MMP-3, -8 and -9) have recently been classified as antitargets due to the key role they play in normal tissue homeostasis. With the precise biological functions of other human MMPs remaining largely unknown, the development of novel chemical and biological methods capable of high-throughput identification and characterization of MMPs has become increasingly urgent. Activity-based profiling (ABP), originally developed by Cravatt et al.,2 is one such technique that has recently been adapted for the study of metalloproteases including MMPs.3 ABP works by selectively targeting enzymes of choice from a crude proteome, in an activity-based manner, using the so-called activity- and affinity-based small molecule probes. In the case of MMPs, small molecule probes possessing a) a hydroxamic acid recognition moiety known to chelate to the active-site zinc of MMPs (as well as other zinc-containing proteins such as thermolysin and collagenase), b) a photolabile group (usually benzophenone or diazirine) capable of covalent crosslinking to the target enzyme, and c) a fluorescent/affinity tag for easy visualization/isolation of the cross-linked enzyme, have been successfully documented.4 Furthermore, in cases where a repertoire of probes having different recognition elements are available, one can obtain the “activity-based fingerprint” of the target enzyme, which reveals not only affinity, but more importantly specificity, of the enzyme/probe complex.5 Most activity-based profiling experiments have been routinely carrried out in gel-based experiments. Recent extension of ABP into a protein microarray has promised even higher throughput, as well as miniaturization, in future enzyme assays.5,6 However, no report has thus far documented the application of affinity-based probes (such as ones that target MMPs4) on a protein microarray.
As shown in Scheme 1, our previously reported first-generation MMP probes were peptides containing a C-terminal hydroxamic acid.4b Despite simplicity in their chemical synthesis, these so-called “left-handed” probes suffer a major drawback in that they bind to MMPs with relatively low affinity, making them less suitable for sensitive detection of MMPs from a complex proteome.1,2 In this Communication, we report 1) the synthesis of second-generation “right-handed” probes aided by “click chemistry”, 2) their application in gel-based activity-based fingerprinting of numerous metalloproteases (including MMPs), and 3) the related preliminary microarray-based experiments which, for the first time, demonstrate that affinity-based probes are indeed compatible with protein microarrays for potential enzyme profiling experiments.
 | ||
| Scheme 1 General structures of the 1st and 2nd generation MMP probes. | ||
“Click Chemistry” is a concept originally introduced by Sharpless et al. which refers to several classes of chemical transformations that share a number of important properties including very high reaction efficiency (in both conversion and selectivity) under mild conditions, and a simple workup.7 The Cu(I)-catalyzed 1,3-cycloaddition between an azide and an alkyne is one such reaction that has recently been explored in the discovery of drugs and materials. Herein, we take advantage of the “click chemistry” between an alkyne-derivatized succinyl hydroxamate warhead and an azide-containing trifunctional tag for the highly modular and facile synthesis of a total of 12 different activity-based probes against MMPs, which are otherwise challenging to make synthetically (Scheme 2).
 | ||
| Scheme 2 General structures of the 12 MMP probes used in this work. Val (A); Leu (B); Ile (C); Phe (D); Long-Phe (E); Cyclohexyl (F); Cyclopentyl (G); O–Ph (H); Long-OH (I); Lys (J); Asp (K); Sulfone (L). | ||
It is noted that, while our manuscript was in preparation, Cravatt et al. reported a similar MMP-targeting, succinyl hydroxamate library, in which “click chemistry” was used to facilitate cell permeability, rather than chemical synthesis, of the probes per se.8
Of the twelve succinyl hydroxamates, eight contain various alkyl and aromatic hydrophobic side chains (A to H in Scheme 2). They were synthesized based on previously reported procedures,9 details of which will be published elsewhere. The remaining four hydroxamates (I to L) introduce side chains possessing acidic and basic groups, as well as hydrophilic groups with the property of hydrogen-bonding, at the P1′ position, thus are able to form favorable electrostatic interactions with different S1′ binding pockets of MMPs. The four corresponding precursors, in suitably protected form as shown in Fig. 1, were accordingly prepared. The acid-labile protecting groups in 1–4 were chosen such that the warheads are compatible with standard solid-phase Fmoc chemistry, allowing them to be used in future for large-scale synthesis of probe libraries.
 | ||
| Fig. 1 Structures of four new hydrophilic succinyl warheads. | ||
Detailed synthesis of the above four warheads is described in the ESI. We took into consideration that the synthetic strategy needs to be general, enabling the facile incorporation of a wide variety of P1′ side chains and, when necessary, with precise stereospecific control over the chiral center located in the warhead. This was accomplished by standard enolate chemistry coupled with Evans' oxazolidinone auxiliary, with different side chains being introduced from their corresponding alkyl bromides. The hydroxamate was protected with a trityl group, ensuring its compatibility with standard Fmoc peptide chemistry/TFA cleavage procedures. The four warheads were synthesized in 5–8 steps from readily available starting materials, with good to excellent yields in almost all steps. With the sulfone-containing warhead, i.e.4, a base-labile protection group was avoided in order to minimize the risk of β-elimination side reaction caused by the release of the sulfone. With all twelve warheads in hand, propargylamine was subsequently used to install the alkyne handle. Prior to or upon TFA treatments, the resulting warheads (in either protected or free form, respectively) were assembled to the azide-containing trifunctional molecule (which contains benzophenone as the photolabile group and rhodamine as the fluorescent tag; see ESI) using “click chemistry”, furnishing the twelve desired MMP probes as shown in Scheme 2. The robustness of “click chemistry” was once again proven to be the key to successful synthesis of our final probes, in that all twelve probes were quantitatively assembled with no trace of side products.
We next sought to fingerprint different metalloproteases including MMPs with our probes. Protein fingerprinting, as its name suggests, is a distinctive pattern generated against a panel of focused small molecule probes, which reflects the protein's catalytic activity or binding property. Since our probes were designed based on known inhibitors of metalloproteases, the resulting inhibitor-based fingerprint not only offers invaluable information for decoding the enzymes' physiological roles, but also facilitates the discovery of potent and selective inhibitors as potential drugs. A diverse group of different classes of metalloproteases were chosen in our experiment, such that they highlight the potential of the strategy not only in distinguishing between both close and distant members, but also in potential identification and characterization of various disease-related enzymes, i.e. MMPs and anthrax lethal factor. In addition, carbonic anhydrase, a well-known zinc-binding protein (but not a metalloprotease) was also tested. Fig. 2 displays the results obtained using gel-based fingerprinting with the probe library. Notably, we were able to produce distinct and reproducible fingerprints for each of these proteins, thus providing a unique capability of identifying and classifying these proteins according to their labeling profiles. Generally the Lys and Ile probes showed the greatest degree of labeling and seemed to strongly label nearly all the enzymes tested. The other probes were however more discerning in their labeling patterns. It was observed that the strongest labeling for MMP-3 was that of Long-OH and Lys probes. This agrees well with the known long hydrophobic pocket of MMP-3 that has been previously reported to bind designed inhibitors with such long hydrophilic scaffolds.1 MT-1 MMP also shares a similar long pocket to MMP-3 and is observed to possess greater affinity to the Long-Phe as well as the Lys probe relative to the other probe scaffolds. The short S1′ pocket of MMP-7 was shown to accommodate both the Val and Phe probes. The Asp and Sulfone probes showed the weakest labeling with most of the enzymes, indicating these moieties are generally unfavored for most of the metalloproteases tested. Overall the fingerprints enabled different enzymes to be classified according to their similarity. MMP-3 gave a distinct profile compared to the other enzymes screened. The labeling pattern of carbonic anhydrase was similar to that of MMP-7. Both anthrax lethal factor and MT-1 MMP show strong selective labeling with one of the probe library set, namely Ile and Lys respectively. Importantly the panel of probes we have designed enables sufficient coverage for one enzyme to be distinguished from the next. We have further shown the fingerprints obtained are activity-based (that is, dependent upon the enzyme catalytic activity), and may be carried out with enzymes present in a crude proteome mixture (that is, in the presence of other unrelated proteins) (see ESI).
 | ||
| Fig. 2 Fingerprints of 12 probes against 7 metalloenzymes. Strongest relative labeling is visualized in red according to the scale shown. The fingerprints were further hierarchically clustered according to their labeling profiles (see ESI). | ||
We next tested the feasibility of these probes to be used in a protein microarray for potential high-throughput discovery of metalloproteases. Previously, only activity-based, and not affinity-based, probes have been shown to detect enzymes immobilized in a protein microarray.5a,6 Five different enzymes, comprising three metalloproteases (i.e. collagenase, thermolysin and anthrax LF), one serine protease (i.e. β-chymotrypsin) and carbonic anhydrase, were spotted in triplicate on a glass slide, and subsequently screened with the Leu probe (Fig. 3). Results indicate that the probe was in general able to distinguish metalloenzyme activity over other non-metalloenzyme activities, in most cases generating positive fluorescence signals only with metalloproteases (e.g. collagenase and thermolysin), as well as carbonic anhydrase (a zinc-binding enzyme), but not with β-chymotrypsin. Despite several attempts, we were unable to detect the fluorescence labeling of anthrax LF, as well as several MMPs (data not shown), on the microarray. As these proteins were only available from commercial sources in very low stock concentrations, we attributed our failure to the less-than-optimal immobilization of the proteins. Work is in progress to confirm this by recombinantly expressing these proteins in sufficient quantity/concentration for protein microarray fabrication, and results will be reported in due course.
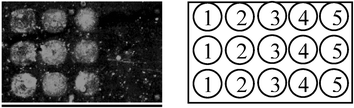 | ||
| Fig. 3 Protein microarray of various metalloenzymes screened by the Leu probe. Five different proteins were spotted in triplicate: 1. carbonic anhydrase (300 µg/ml); 2. collagenase (300 µg/ml); 3. thermolysin (300 µg/ml); 4. anthrax LF (6 µg/ml); 5. β-chymotrypsin (300 µg/ml). | ||
In conclusion, we have used “click chemistry” to successfully synthesize a second-generation library of metalloprotease probes containing succinyl warheads with a variety of P1′ functionalities. With these probes, we have been able to generate unique activity-based fingerprints against various metalloproteases including MMPs and other therapeutically important enzymes such as anthrax LF. Such fingerprinting strategies may lead to future identification and characterization of new MMPs, and the development of potential potent and selective inhibitors. We have also for the first time shown that affinity-based probes may be equally amenable for high-throughput screening of metalloprotease activities in a protein microarray.
Funding support was provided by the National University of Singapore (NUS) and the Agency for Science, Technology and Research (A*STAR) of Singapore.
Notes and references
- C. M. Overall and O. Kleifeld, Nat. Rev. Cancer, 2006, 6, 227–239 CrossRef CAS.
- Y. Liu, M. P. Patricelli and B. F. Cravatt, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 14694–14699 CrossRef CAS.
- For a recent review, see: A. Saghatelian and B. F. Cravatt, Nat. Chem. Biol., 2005, 1, 130–142 Search PubMed.
- (a) A. Saghatelian, N. Jessani, A. Joseph, M. Humphrey and B. F. Cravatt, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 10000–10005 CrossRef CAS; (b) E. W. S. Chan, S. Chattopadhaya, R. C. Panicker, X. Huang and S. Q. Yao, J. Am. Chem. Soc., 2004, 126, 14435–14446 CrossRef CAS.
- (a) R. Srinivasan, X. Huang, S. L. Ng and S. Q. Yao, ChemBioChem, 2006, 7, 32–36 CrossRef; (b) D. C. Greenbaum, W. D. Arnold, F. Lu, L. Hayrapetian, A. Baruch, J. Krumrine, S. Toba, K. Chehade, D. Bromme, I. D. Kuntz and M. Bogyo, Chem. Biol., 2002, 9, 1085–1094 CrossRef CAS.
- (a) G. Y. J. Chen, M. Uttamchandani, Q. Zhu, G. Wang and S. Q. Yao, ChemBioChem, 2003, 4, 336–339 CrossRef CAS; (b) D. P. Funeriu, J. Eppinger, L. Denizot, M. Miyake and J. Miyake, Nat. Biotechnol., 2005, 23, 622–627 CrossRef CAS.
- H. C. Kolb and K. B. Sharpless, Drug Discovery Today, 2003, 8, 1128–1137 CrossRef CAS.
- S. A. Sieber, S. Niessen, H. S. Hoover and B. F. Cravatt, Nat. Chem. Biol., 2006, 2, 274–281 CrossRef CAS.
- J. Wang, M. Uttamchandani, L. P. Sun and S. Q. Yao, Chem. Commun., 2006, 717–719 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Procedures for the synthesis and characterization of inhibitors, microarray and microplate experiments, as well as kinetic experiments and computer modeling. See DOI: 10.1039/b609446e |
| This journal is © The Royal Society of Chemistry 2006 |
