PAMAM dendrimers for efficient siRNA delivery and potent gene silencing†
Jiehua
Zhou
a,
Jiangyu
Wu
ab,
Nadia
Hafdi
c,
Jean-Paul
Behr
d,
Patrick
Erbacher
c and
Ling
Peng
*ab
aCollege of Chemistry and Molecular Sciences, Wuhan University, Wuhan, 430072, China
bDépartement de Chimie, CNRS UMR 6114, 163 avenue de Luminy, 13288 Marseille cedex 09, France. E-mail: ling.peng@univmed.fr; Fax: 00 33 4 91 82 93 01; Tel: 00 33 4 91 82 91 54
cPolyplus-transfection SA, Bioparc, Boulevard S. Brandt, BP90018, 67401 Illkirch, France. E-mail: perbacher@polyplus-transfection.com
dLaboratoire de Chimie Génétique, Faculté de Pharmacie, CNRS UMR7514, 67401 Illkirch, France
First published on 10th May 2006
Abstract
Genuine, nondegraded PAMAM dendrimers self-assemble with siRNA into nanoscale particles that are efficient for siRNA delivery and induce potent endogenous gene silencing.
Short double-stranded RNAs, which are known as short interfering RNA (siRNA), can be used to specifically down-regulate expression of the targeted gene in a process known as RNA interference (RNAi).1 RNAi has rapidly become a popular tool for functional genomics and target gene validation, and holds great promise for therapeutic applications as well.1 However, the success of gene silencing applications based on the use of synthetic siRNA critically depends on efficient intracellular delivery. There is therefore an urgent need for safe and efficient siRNA delivery systems.2–6
Polycationic dendrimers such as poly(amidoamine) (PAMAM) dendrimers7,8 are known to be efficient DNA delivery systems.9 These dendrimers bear primary amine groups on their surface, while having tertiary amine groups inside. The primary amine groups participate in DNA binding, compact DNA into nanoscale particles and promote the cellular uptake of DNA, while the buried tertiary amino groups act as a proton sponge in endosomes and enhance the release of DNA into the cytoplasm. Moreover, partially degraded PAMAM dendrimers were reported to have more flexible structures than intact dendrimers and therefore to interact more efficiently with DNA.10 Their excellent DNA delivery properties have been confirmed now. All these features of dendrimers could also be used for RNA delivery, although very few efforts have been made so far along these lines.11 In our ongoing project focusing on RNA targeting and RNA delivery, we are interested in developing flexible polycationic PAMAM dendrimers for siRNA delivery purposes. The advantages of using dendrimers over other delivery systems are their controllable molecular structure and size, high chemical and structural homogeneity, high ligand and functionality density etc., which will allow us to fine-tune dendrimer properties for the delivery purpose.
We recently reported that PAMAM dendrimers showed strong binding affinity for RNA molecules.12,13 Dendrimers with the triethanolamine as core have the branching units starting 10 successive bonds from the center amine (Scheme 1). Therefore, these dendrimers are expected to have less densely packed branching units and end groups than the commercially available PAMAM dendrimers with NH3 or ethylenediamine as core, where the branching starts immediately at the center amine of the core. We have shown recently that lower-generation dendrimers belonging to this family showed strong interactions with group I intron ribozymes.12 Here, we report that higher-generation dendrimers belonging to this family can efficiently deliver siRNA and thus induce gene silencing in cell culture.
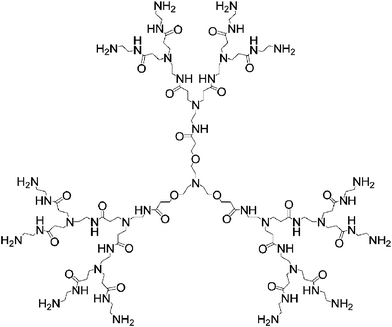 | ||
| Scheme 1 Chemical structure of PAMAM dendrimers with triethanolamine as core. For clarity, only G2 is drawn. | ||
Dendrimers used in this study are referred as Gn (n: dendrimer generation number, Scheme 1). From generation 1 to 7, the corresponding dendrimer has, respectively, 6, 12, 24, 48, 96, 192 and 384 end amine groups. We first studied the ability of dendrimers to form self-assembled complexes with siRNA by performing agarose gel electrophoresis. Dendrimers with amine end groups were able to almost completely retard siRNA in the gels at N/P ratios > 2.5 (Fig. 1A, N/P is the ratio of the total number of dendrimer end amine groups and the total number of siRNA phosphate groups). No gel retardation was observed with dendrimers having ester end groups, even at N/P charge ratios up to 20 (data not shown). These findings show that strong binding occurred between siRNA molecules and the dendrimers with amine end groups, indicating that an electrostatic complex is formed between the positively charged amine end groups at the dendrimer surface and the negatively charged phosphate groups of siRNA. In addition, formation of the siRNA–dendrimer complex depends on the dendrimer generation (data not shown). The higher the generation of the dendrimers, the stronger the interactions between the dendrimer and the RNA, and therefore the more stable the complex formed. This is similar to what has been observed in the case of DNA–dendrimer9 and ribozyme–dendrimer complexes.12,13
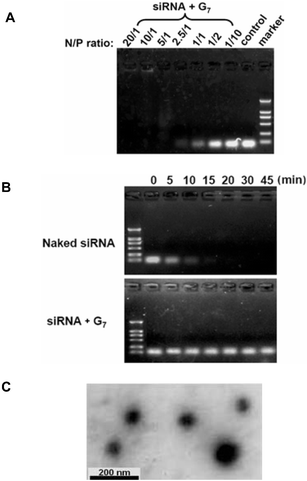 | ||
| Fig. 1 (A) Agarose gel analysis of the siRNA–dendrimer complexes. Migration of the siRNA/G7 complexes at charge ratios N/P varying from 1/10 to 20/1 in Tris–HCl buffer solution (pH 7.6). (B) Dendrimer G7 is able to prevent siRNA from RNase A degradation. Naked siRNA and siRNA/G7 complex (N/P 2.5/1) were incubated in the presence of 0.01 μg/μL RNase A for 0, 5, 10, 15, 20, 30 and 45 min at 37 °C and then agarose gel electrophoresis was performed. (C) TEM imaging of the siRNA/G7 complex prepared in Tris–HCl buffer, pH 7.6. | ||
The siRNA–dendrimer complexes are very stable. Disruption of these complexes required strong ionic detergents such as SDS (data not shown), and was not observed under physiological conditions. Moreover, the siRNA–dendrimer complexes showed considerable resistance to RNase degradation, contrary to naked siRNA (Fig. 1B). This is in agreement with results obtained from transmission electron microscopy (Fig. 1C), which showed that the siRNA and dendrimer formed compacted, spherical, nanosize particles (ca. 60–110 nm in diameter). Therefore, these dendrimers self-assemble with siRNA into nanosized particles, similar to what has been observed with DNA–dendrimer9 and ribozyme–dendrimer complexes.13
The ability of siRNA–dendrimer complexes to deliver siRNA to cells was tested using a model of endogenous gene knockdown. For these assays, A549Luc cells stably expressing the GL3 luciferase gene were used. Efficient gene silencing was observed with the specific GL3Luc siRNA–G7 complex (Fig. 2A), while neither the naked siRNA (data not shown) nor the nonspecific GL2Luc siRNA–G7 complex showed any gene silencing effect (Fig. 2A). These results are a consequence of efficient dendrimer-mediated siRNA delivery.
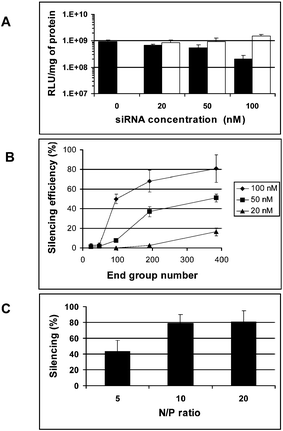 | ||
| Fig. 2 Endogenous silencing efficiency in A549Luc cells stably expressing GL3Luc with (A) dendrimer G7 and, respectively, the specific siRNA (GL3Luc siRNA, black) and the non-specific siRNA (GL2Luc siRNA, white), at various concentrations; (B) dendrimers of various generations and the GL3Luc siRNA with various concentrations at N/P ratio of 10; (C) denrimer G7 and the GL3Luc siRNA at various N/P ratios. | ||
Moreover, the gene silencing ability of the siRNA–dendrimer complex was dependent on the dendrimer generation (Fig. 2B), the N/P ratio (Fig. 2C), and the siRNA concentration (Fig. 2A and Fig. 2B). The siRNA–dendrimer complex showed generation-dependent gene silencing behavior: the higher the dendrimer generation, the more efficient the gene silencing was found to be. In a recent report, only weak gene silencing effect was observed when using a conjugated dendrimer of generation 5.11 In our system, the best gene silencing results were obtained with G7 at an N/P ratio of 10–20 and siRNA concentration of 100 nM, where more than ca. 80% gene silencing was observed (Fig. 2). This correlates well with the results obtained on complex formation between dendrimers and siRNA, which showed that the siRNA complexing ability of the dendrimer increased with the generation number, due to the increasingly strong interactions occurring between dendrimer and siRNA via cooperative interactions. The fact that the gene silencing efficiency was greater at higher N/P ratio may be attributed to both the formation of stronger siRNA–dendrimer complexes at neutral pH and the efficient internalization of the siRNA molecules into the cytoplasm through buffering of the endosomal cavity due to the relative high buffer capacity of dendrimer G7, which has the continuous lower pKa values (Fig. 3), as occurs in the case of PEI14 used for DNA transfection. PEI seems not to be an efficient vector for siRNA delivery,15 yet there are also reports on siRNA-mediated gene silencing using PEI.16
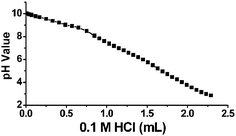 | ||
| Fig. 3 Potentiometric pH titration curve of dendrimer G7. | ||
We also followed the kinetics of gene silencing with G7 (Fig. 4). The silencing efficiency was stable up to 48 h and remained at a level of around 50% even after 72 h, indicating slow siRNA release over an extended period of time. This finding suggests that our dendrimer is an effective siRNA delivery system and that siRNA–dendrimer complexes are able to produce efficient and long-term gene silencing.
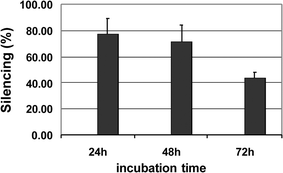 | ||
| Fig. 4 Kinetic of gene silencing with dendrimer G7 and GL3Luc siRNA. | ||
We finally examined the toxicity of dendrimers using the MTT assay (Fig. 5). The delivery step was relatively safe for the cells although some toxicity was observed for the highest siRNA concentration.
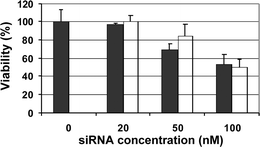 | ||
| Fig. 5 Cell viability determined by MTT assay 24 h after the transfection in A549 cells with dendrimer G7 and siRNA (GL3Luc siRNA in black and GL2Luc siRNA in white) at N/P ratio of 10. | ||
In conclusion, we report here on an effective siRNA delivery system based on structure flexible polycationic PAMAM dendrimers. These genuine, nondegraded dendrimers condense siRNA into nanoscale particles. They protect siRNA from enzymatic degradation and achieve substantial release of siRNA over an extended period of time for efficient gene silencing. Taken together, these findings suggest that flexible polycationic dendrimers may have great potential for in vitro (functional genomics) and possibly in vivo (therapeutic) applications. The possibility of closely controlling the size, shape and surface chemistry of dendrimers should give us a unique opportunity for fine-tuning dendrimer properties17 with the view to improving the delivery efficiency and lowering the cell toxicity. One further direction is to use dendrimers for cell-specific delivery of siRNA. For this purpose, dendrimers are of special interest because one can develop fan-shaped multi-functional compounds with one end attached to a cell-specific ligand and the other end for dendrimer promotion18 and siRNA binding. Studies in these directions are currently underway at our laboratories.‡
This work was supported by the Ministry of Science and Technology of China, National Science Foundation of China, Cheung-Kong Scholar Foundation, Wuhan University, the Association Française contre les Myopathies and the Centre National de la Recherche Scientifique. We are grateful to Dr Jean Lorquin for his help in HPLC analysis. ZJH, WJY and NH contributed equally to this work.
Notes and references
- R. S. Dave and R. J. Pomerantz, Rev. Med. Virol., 2003, 3, 373–385 CrossRef; A. Eccleston and A. K. Eggleston, Nature, 2004, 431(Insight – RNA Interference), 337–378 CrossRef CAS; G. Riddihough, Science, 2005, 309(Special issue – Mapping RNA Form & Function), 1507–1533 CrossRef CAS.
- K. Itaka, N. Kanayama, N. Nishiyama, W.-D. Jang, Y. Yamasaki, K. Nakamura, H. Kawaguchi and K. Kataoka, J. Am. Chem. Soc., 2004, 126, 13612–13613 CrossRef CAS.
- Y. Minakuchi, F. Takeshita, N. Kosaka, H. Sasaki, Y. Yamamoto, M. Kouno, K. Honma, S. Nagahara, K. Hanai, A. Sano, T. Kato, M. Terada and T. Ochiya, Nucleic Acids Res., 2004, 32, e109 CrossRef.
- S. Spagnou, A. D. Miller and M. Keller, Biochemistry, 2004, 43, 13348–13356 CrossRef CAS.
- B. Dalby, S. Cates, A. Harris, E. C. Ohki, M. L. Tilkins, P. J. Price and V. C. Ciccarone, Method, 2004, 33, 95–103 Search PubMed.
- N. Unnamalai, B. G. Kang and W. S. Lee, FEBS Lett., 2004, 566, 307–310 CrossRef CAS.
- J. M. J. Fréchet and D. A. Tomalia, Dendrimers and other dendritic polymers, Wiley, Chichester, 2001 Search PubMed.
- D. A. Tomalia, H. Baker, J. Dewald, M. Hall, G. Kallos, S. Martin, J. Roeck, J. Ryder and P. Smith, Polym. J., 1985, 17, 117–132 CAS.
- J. Haensler and F. C. Szoka, Bioconjugate Chem., 1993, 4, 372 CrossRef CAS; J. F. Kukowska-Latallo, A. U. Bielinska, J. Johnson, R. Spindler, D. A. Tomalia and J. R. Baker, Jr., Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 4897–4902 CrossRef CAS.
- M. X. Tang, C. T. Redemann and F. C. Szoka, Jr., Bioconjugate Chem., 1996, 7, 703–714 CrossRef CAS.
- H. Kang, R. Delong, M. H. Fisher and R. L. Juliano, Pharm. Res., 2005, 22, 2099–2106 CrossRef CAS.
- J. Y. Wu, J. H. Zhou, F. Q. Qu, P. H. Bao, Y. Zhang and L. Peng, Chem. Commun., 2005, 313–315 RSC.
- J. H. Zhou, J. Y. Wu, X. X. Liu, F. Q. Qu, M. Xiao, Y. Zhang, L. Charles, C.-C. Zhang and L. Peng, Org. Biomol. Chem., 2006, 4, 581–586 RSC.
- O. Boussif, F. Lezoualc'h, M. A. Zanta, M. D. Mergny, D. Scherman, B. Demeneix and J. P. Behr, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 7297–7301 CAS.
- B. Li, Q. Tang, D. Cheng, C. Qin, F. Y. Xie, Q. Wei, J. Xu, Y. Liu, B. Zheng, M. C. Woodle, N. Zhong and P. Y. Lu, Nat. Med., 2005, 11, 944–951 CAS; P. Erbacher and J. P. Behr, unpublished results.
- Q. Ge, L. Filip, A. Bai, T. Nguyen, H. N. Eisen and J. S. Chen, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 8676–8681 CrossRef CAS; B. Urban-Klein, S. Werth, S. Abuharbeid, F. Czubayko and A. Aigner, Gene Ther., 2005, 12, 461–466 CrossRef CAS.
- C. C. Lee, J. A. MacKay, J. M. Fréchet and F. C. Szoka, Nat. Biotechnol., 2005, 23, 1517–1526 CrossRef CAS and references therein.
- K. C. Wood, S. R. Little, R. Langer and P. T. Hammond, Angew. Chem., Int. Ed., 2005, 44, 6704–6708 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: detailed materials and methods section. See DOI: 10.1039/b601381c |
| ‡ Detailed materials and methods section. This material is available in ESI†. |
| This journal is © The Royal Society of Chemistry 2006 |
