Synthetic mimics of mammalian cell surface receptors: prosthetic molecules that augment living cells
Blake R.
Peterson
*
Department of Chemistry, The Pennsylvania State University, 104 Chemistry Building, University Park, PA 16802, USA. E-mail: brpeters@chem.psu.edu
First published on 8th September 2005
Abstract
Specific receptors on the surface of mammalian cells actively internalize cell-impermeable ligands by receptor-mediated endocytosis. To mimic these internalizing receptors, my laboratory is studying artificial cell surface receptors that comprise N-alkyl derivatives of 3β-cholesterylamine linked to motifs that bind cell-impermeable ligands. When added to living mammalian cells, these synthetic receptors insert into cellular plasma membranes, project ligand-binding small molecules or peptides from the cell surface, and enable living cells to internalize targeted proteins and other cell-impermeable compounds. These artificial receptors mimic their natural counterparts by rapidly cycling between plasma membranes and intracellular endosomes, associating with proposed cholesterol and sphingolipid-rich lipid raft membrane microdomains, and delivering ligands to late endosomes/lysosomes. This “synthetic receptor targeting” strategy is briefly reviewed here and contrasted with other related cellular delivery systems. Potential applications of artificial cell surface receptors as molecular probes, agents for cellular targeting, tools for drug delivery, and methods for ligand depletion are discussed. The construction of synthetic receptors as prosthetic molecules, designed to seamlessly augment the molecular machinery of living cells, represents an exciting new frontier in the fields of bioorganic chemistry and chemical biology.
 Blake R. Peterson Blake R. Peterson | Blake Peterson was born on March 28, 1968 and raised in Reno, Nevada. After receiving a BS degree in chemistry in 1990 from the University of Nevada, Reno, he pursued graduate studies in chemistry with Prof. François Diederich at both UCLA (1990–1992) and the ETH Zürich (1992–1994). Following completion of his UCLA PhD in 1994, he joined the laboratory of Prof. Gregory Verdine in the Dept. of Chemistry and Chemical Biology at Harvard University as a Damon Runyon-Walter Winchell Cancer Research Foundation Postdoctoral Fellow. He joined the faculty in the Dept. of Chemistry at Penn State University as an Assistant Professor in 1998 and was promoted to Associate Professor with tenure in 2004. He was named a research scholar of the American Cancer Society in 2003 and was the recipient of a Camille Dreyfus Teacher Scholar Award in 2004. His laboratory focuses on the synthesis of small molecules that control and probe cellular biology. |
Introduction
Mammalian cells sport a vast array of structurally divergent receptors on their surfaces. Some of these receptors are large transmembrane proteins over one hundred thousand dalton in molecular weight that play key roles in cellular signaling or nutrient uptake. Other much smaller receptors include glycolipids, typically less than two thousand dalton, involved in the cellular processes of endocytosis and adhesion. A defining feature of these diverse biomolecules is a ligand-binding motif that projects from the cell surface and engages a specific molecule in the aqueous environment. These motifs are typically attached to the cell by either protein alpha helices that span the plasma membrane or by covalently linked lipids that insert into the outer leaflet of the membrane's lipid bilayer. Transmembrane segments of cell surface proteins often protrude into the cytoplasm and interact with intracellular proteins. This bridge to the interior of the cell enables ligands that bind on the cell surface to transmit signals to effector molecules in the cellular cytoplasm. These effectors include enzymes involved in cellular signaling and components of the molecular machinery controlling the ligand uptake process of endocytosis. Receptor-mediated endocytosis is a major mechanism by which cells consume nutrients available in the environment and terminate signals initiated by extracellular proteins. This process is also exploited by opportunistic proteins and pathogens to invade living cells.1The LDL receptor: a prototypical macromolecular cell surface receptor
One of the most extensively characterized internalizing cell surface receptors is the low-density lipoprotein (LDL) receptor.2–4 As illustrated in Fig. 1, this large single-pass transmembrane protein specializes in the uptake of cholesterol-laden LDL particles. By recognizing the Apo-B protein component of LDL, the LDL receptor enables cells to internalize exogenous cholesterol, a key building block required for the biosynthesis of steroid hormones, bile acids, and cellular plasma membranes. For this reason, the LDL receptor is often overexpressed on rapidly proliferating cancer cells in demand of high levels of membrane biosynthesis, providing a target for the selective delivery of anticancer and tumor imaging agents.5,6 As shown in Fig. 2, receptor-mediated endocytosis of LDL is initiated by clustering of receptor/ligand complexes on the cell surface in pits coated by the intracellular protein clathrin. Clathrin controls the endocytosis of many cell surface receptors, and this membrane-associated protein interacts either directly or indirectly with the intracellular domain of the LDL receptor. Clathrin-coated pits containing LDL-bound LDL receptors invaginate and pinch off to form intracellular endocytic vesicles. These internalized vesicles are acidified by the activation of proton pumps and fuse in the cytoplasm to form larger acidic (pH ∼ 6) endosomes. Receptors dissociate from ligands in endosomes, and the free receptor cycles back to the cell surface, allowing the LDL receptor to be reused up to several hundred times during its ∼20 h lifespan. Endosomes containing free LDL fuse with lysosomes, more acidic (pH ∼ 5) organelles containing hydrolytic enzymes that liberate needed cholesterol and amino acid nutrients for delivery into the cytoplasm.2,3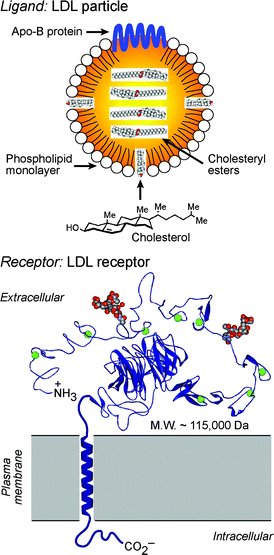 | ||
| Fig. 1 Structural depictions of the LDL particle (top) and its cell surface receptor (bottom). The extracellular domain of the LDL receptor (shown as the X-ray structure, PDB: 1N7D) binds the Apo-B protein of LDL to initiate receptor-mediated endocytosis of this ligand. | ||
 | ||
| Fig. 2 Endocytosis of LDL (sphere) mediated by the LDL receptor (R). Cellular demand for cholesterol results in upregulation of biosynthesis of the LDL receptor in membranes of the endoplasmic reticulum, and the receptor is secreted through the Golgi complex to the cell surface. Binding of the receptor to LDL in the extracellular environment results in uptake of this ligand by endocytosis, trafficking of LDL to degratory lysosomes, and recycling of the receptor back to the cell surface. | ||
Ganglioside GM1: a small internalizing cell surface receptor
Despite its small size (molecular weight = 1630 Da), ganglioside GM1 (Fig. 3) functions similarly as an internalizing cell surface receptor. This small receptor is targeted by the protein cholera toxin,7 which penetrates into cells by endocytosis after multivalent binding to the glycolipid pentasaccaride headgroup.8–10 However, because ganglioside GM1 is restricted to the outer leaflet of the plasma membrane, the molecular machinery that regulates the endocytosis of the cholera toxin differs in certain respects from that used to internalize LDL. The endocytosis of the cholera toxin, mediated by GM1, has been proposed to involve lipid rafts, plasma membrane subdomains enriched in cholesterol and sphingolipids.11–14 Many proteins covalently or noncovalently associated with cholesterol, sphingolipids such as ganglioside GM1, or saturated lipids of cellular membranes, are thought to associate with lipid rafts.15 These membrane subdomains have been proposed to segregate and concentrate membrane proteins, regulate the activation of specific signal transduction pathways,16 and control the endocytosis of specific receptors.17 In certain cell lines, particularly those with high expression levels of ganglioside GM1,18 endocytosis of the cholera toxin is thought to involve lipid raft-dependent pathways.19 However, in enterocytes and neurons, endocytosis of the cholera toxin is thought to predominantly involve clathrin.20 Multiple, simultaneous, mechanisms of endocytosis of this toxin have also been observed.21,22 Yet, the mechanism that couples recognition of the glycolipid on the outer leaflet of the membrane to the clathrin machinery on the inner leaflet remains to be defined. The bound cholera toxin is initially delivered by GM1 into endosomes, but this protein bypasses lysosomes, traffics into the Golgi complex and endoplasmic reticulum, and escapes into the cytosol, where it functions as a catalytic toxin. Many protein toxins and viruses exploit receptor-mediated endocytosis involving lipid rafts or clathrin to access the interior of living cells.23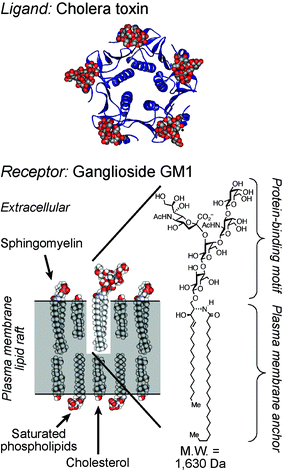 | ||
| Fig. 3 Structures of the opportunistic ligand cholera toxin (top) and its small cell surface receptor ganglioside GM1 in a model lipid raft (bottom). The X-ray structure of the cholera toxin B-subunit is shown bound to GM1 oligosaccarides (PDB: 3CHB). | ||
Due to the efficiency of receptor-mediated endocytosis as a delivery mechanism, cell surface receptors involved in this process have been extensively targeted to enable cellular uptake of poorly permeable molecules. Ligands of internalizing receptors have been linked to drugs, molecular probes, and macromolecules to deliver these conjugates into cells.24 Receptors targeted in this way include LDL receptors,6 glycosylphosphatidylinositol (GPI) lipid-anchored folate receptors,25 and transferrin receptors,26 all of which tend to be overexpressed on rapidly proliferating cells. Low expression of these receptors provides a common mechanism of resistance to these types of targeted therapeutics.
Artificial cell surface receptors: tools for synthetic receptor targeting
My laboratory is investigating synthetic compounds designed to mimic mammalian cell surface receptors. These receptor mimics are designed to enable receptor-mediated endocytosis of impermeable ligands that do not bind natural receptors expressed on cell surfaces. These artificial receptors comprise the non-natural membrane anchor N-alkyl-3β-cholesterylamine linked to binding motifs for proteins and other poorly permeable compounds. As shown in Fig. 4, we have linked this membrane anchor to fluorophores (receptors 1, 7, and 8),27,28 the dinitrophenyl hapten (receptor 6),28 biotin (receptor 2),29 and peptides (receptors 3, 4, and 5)30 as protein binding motifs. When added to growth media containing mammalian cells, these compounds rapidly become incorporated into cellular plasma membranes. The half-lives of these artificial receptors on the surface of living cells, as detected with a fluorescent ligand (e.g.t1/2(6) ∼ 20 h),28 are comparable to those of GPI-linked folate receptors (t1/2 ∼ 24 h).31 Mammalian cells treated with these synthetic receptors gain the capacity to endocytose normally impermeable protein ligands, and these ligands are typically delivered to late endosomes and lysosomes.27–30 Because of these functional similarities between our synthetic receptors (Fig. 4) and other internalizing cell surface receptors, we termed this delivery approach “synthetic receptor targeting” (Fig. 5).28,30 | ||
| Fig. 4 Structures of previously reported synthetic cell surface receptors derived from N-alkyl-3β-cholesterylamine. Ligands: antifluorescein IgG (receptors 1 and 8), streptavidin (receptors 2 and 5), anti-hemagglutinin tag IgG (receptor 3), anti-flag tag IgG (receptor 4), and anti-dinitrophenyl IgG (receptors 6 and 7). | ||
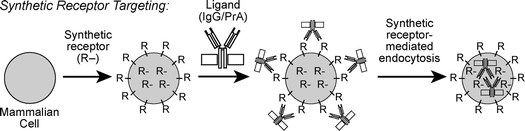 | ||
| Fig. 5 The synthetic receptor targeting approach for enhancing the cellular uptake of impermeable ligands. Living mammalian cells are treated with synthetic mimics of cell surface receptors (R). These receptors become incorporated in cellular plasma membranes, enabling the cellular uptake of cognate ligands such as macromolecular antibodies (IgG) bound to bacterial protein A (PrA) by synthetic receptor-mediated endocytosis. The receptors in the center of the cell represent the population in intracellular endosomes in dynamic exchange with receptors on the cell surface. | ||
The cellular uptake of ligands mediated by receptors derived from N-alkyl-3β-cholesterylamines appears to mimic endocytic cellular penetration mechanisms employed by certain protein toxins and viruses.23 These mechanisms are highly complex at the molecular level and appear to involve clathrin-mediated endocytosis, raft-mediated endocytosis, or a combination thereof, depending on the cell type. Sucrose density gradient ultracentrifugation analysis of cells treated with synthetic receptor 6 and fluorescent anti-dinitrophenyl (anti-DNP) IgG ligand revealed that this ligand co-fractionates with cholera toxin in a putative low-density lipid raft fraction of the plasma membrane.28 These and other results suggest significant similarities between the initial steps of uptake of cholera toxin, mediated by ganglioside GM1, and the uptake of ligands mediated by synthetic receptors such as 1–8.
In the absence of a ligand, many N-alkyl-3β-cholesterylamine derivatives rapidly and constitutively cycle between the cell surface and endosomes. For example, receptor 8 makes a round trip between these compartments in approximately 10 min.28 The fluorescent sphingolipid C6-NBD-sphingomyelin32 and the macromolecular LDL receptor3 exhibit similar kinetics of constitutive plasma membrane recycling.33 However, the internalization of ligands such as anti-DNP IgG mediated by receptor 6 is slower (t1/2 ∼ 95 min)28 than the uptake of LDL by the LDL receptor (t1/2 < 10 min),3 possibly due to the ability of the LDL receptor to interact directly or indirectly with clathrin through its intracellular domain. Despite this slower rate of ligand uptake, treatment of human Jurkat lymphocytes for 1 h with synthetic receptor 6 (10 µM), followed by the washing of cells to remove any unincorporated receptor, and the addition of the ligand for an additional 4 h, enhances the endocytic uptake of fluorescent anti-DNP IgG bound to bacterial Protein A34 by over 200-fold with low toxicity.28 The effectiveness of this delivery system appears to relate in part to the ability of receptor 6 to dissociate from ligands in acidic endosomes and return to the cell surface by plasma membrane recycling for additional rounds of delivery. A simple model of synthetic receptor-mediated endocytosis is shown in Fig. 6.
 | ||
| Fig. 6 A simple model of synthetic receptor-mediated endocytosis. Synthetic receptors embedded in the cellular plasma membrane rapidly cycle between the cell surface and intracellular endosomes. Binding of ligand (IgG) results in association with lipid rafts and uptake of the complex by endocytosis. Dissociation in endosomes frees the receptor to return to the cell surface. The protein ligand is sorted to late endosomes and lysosomes. | ||
Cholesterol is a major component of both mammalian plasma membranes and endosomes involved in plasma membrane recycling.35 Hence endocytic recycling of N-alkyl-3β-cholesterylamine derivatives is likely to relate to mimicry of this critical membrane sterol. However, the appended linker region and protein-binding headgroups of synthetic receptors derived from this cholesterol mimic can substantially affect subcellular trafficking and the localization on the cell surface and on intracellular membranes. Studies of receptors 6 and 7 and related compounds revealed that insertion of β-alanine amino acids into this linker region can substantially increase the number of these receptors on the cell surface compared with the number in intracellular endosomes.28 For example, fluorescence-quenching experiments with receptor 7 (bearing two β-alanine subunits in the linker region) revealed that after treatment of cells for 1 h, 53% of this compound was localized on the plasma membrane, with the other 47% residing in endosomes. Under the same conditions, removal of one β-alanine from the linker reduced the cell surface population to 23%, and removal of both β-alanines resulted in only 3% of the compound on the plasma membrane, with the remainder in endosomes. Not surprisingly, the percentage of receptors on the cell surface is highly correlated with the efficiency of these compounds as mediators of ligand uptake.28
The secondary amine of N-alkyl-3β-cholesterylamines is protonated at physiological pH. This protonation stabilizes association of the membrane anchor with the cell surface and may contribute to efficient plasma membrane recycling. In contrast, structurally related N-acyl derivatives (amide analogues) of 3β-cholesterylamine often differ significantly in membrane trafficking and generally are significantly less effective as synthetic receptors. Unlike 3β-amines that cycle between the cell surface and endosomes, analogous 3β-amides, particularly those bearing uncharged headgroups, tend to localize at internal membranes of the Golgi complex and nuclear membrane of living cells.28 Fluorescent esters of cholesterol similarly localize at these intracellular membranes,36 and N-acyl derivatives of 3β-cholesterylamines presumably mimic the trafficking of natural cholesteryl esters. Although the molecular determinants governing the localization of synthetic receptors on specific membranes of living cells are not yet fully defined, interactions of the membrane anchor and the headgroup with neighboring lipids, proteins, and carbohydrates involved in membrane trafficking is likely to determine the ultimate subcellular destination of these compounds.
Membrane anchors derived from N-alkyl-3β-cholesterylamine can insert into cellular plasma membranes of living mammalian cells and rapidly cycle between the cell surface and endosomes. This plasma membrane recycling enables derivatives linked to motifs that bind proteins and other impermeable ligands to function as artificial cell surface receptors. The linker region of these compounds can be tuned to maximize the fraction of receptor on the cell surface and optimize ligand uptake by these compounds. Artificial cell surface receptors constructed in this way provide unique probes of molecular recognition on cell surfaces and novel agents for the delivery of myriad compounds into mammalian cells.
Other related approaches for the endocytic delivery of impermeable compounds
Several cell surface engineering approaches have been used to enhance the cellular uptake of impermeable molecules.37 One strategy termed “cellular painting” incorporates proteins linked to glycosylphosphatidylinositol (GPI) lipid anchors into cellular plasma membranes.38–42 Cellular painting has primarily been used to study cellular signaling, plasma membrane organization, and immunological responses to modified cell surfaces. However, GPI-linked proteins undergo raft-mediated endocytosis43 and can be used to deliver molecules into cells. For example, Jurkat lymphocytes treated with a GPI-linked variant of an immunoglobulin Fc receptor (FcγRIII) will endocytose an antibody directed against this receptor.44 Structurally related artificial cell surface receptors have also been construced from single-chain antibodies covalently linked to lipids.45 In another related approach, polyethylene glycol (PEG)-linked lipids covalently46 or noncovalently47 attached to proteins have been used to immobilize macromolecules on cell surfaces. These PEG derivatives primarily anchor proteins on the outer leaflet of the plasma membrane, but, in certain cases, they can also promote the endocytosis of protein ligands.The installation of abiotic functional groups into carbohydrates on cell surfaces by metabolic oligosaccaride engineering is another approach to add prosthetic molecules to living cells.48,49 By feeding cells unnatural sugars, cellular metabolism can be harnessed to display bioorthogonal functional groups such as ketones and azides on cell surfaces. Reaction of these ketones with hydrazine derivatives to yield hydrazones,50 as well as the reaction of azides with modified phosphines in the Staudinger ligation,51 can immobilize molecules on the cell surface and promote delivery of proteins such as the toxin ricin.50 This approach has been shown to function in living animals.52
Cell penetrating peptides and proteins (CPPs) represent another approach widely used to deliver molecules into cells both in vitro and in vivo.53–58 CPPs comprise basic segments of HIV Tat and other proteins, polyarginine peptides, and related cationic oligomers. These delivery agents are generally fused or otherwise covalently linked to impermeable proteins and drugs to promote cellular uptake. Mechanisms of cellular uptake of CPPs are thought to be either endocytic or non-endocytic depending on the CPP, the size of its cargo, and the cell type. Some CPPs, particularly linked to a large cargo such as proteins,59 undergo endocytosis mediated by binding to anionic cell surface components such as extracellular heparin sulfates.60
Future directions
Modern techniques of molecular biology provide powerful genetic tools to endow cells with new molecules capable of unique functions. For example, cells transfected with genes encoding artificial cell surface receptors that bind the imaging agent technetium-99 enable tracking of cells in vivo.61 In contrast, the use of synthetic molecules as cellular prostheses, adding new functions such as endocytosis, is much less well developed. However, this chemical approach for controlling biology has the potential to vastly increase the diversity of molecules that can be interfaced with cells, tissues, and organisms.As a chemical approach to endow living cells with the capacity to internalize specific molecules, artificial cell surface receptors can be constructed from the membrane anchor N-alkyl-3β-cholesterylamine. These compounds can exhibit long cellular half-lives, rapidly cycle between the cell surface and intracellular endosomes, and mimic the internalization functions of many mammalian cell surface receptors. Because N-alkyl-3β-cholesterylamine derivatives appear to mimic aspects of the subcellular trafficking of cholesterol, these compounds may provide useful probes of cholesterol-rich membrane subdomains and associated mechanisms that regulate the segregation, localization, and sorting of membrane-bound biomolecules. Synthetic cell surface receptors may also provide novel approaches for cellular targeting and drug delivery. Cellular targeting applications may be facilitated by the structural similarity of these compounds to cholesterol, which may allow packaging of synthetic receptors in LDL or other cholesterol-carrying particles for selective delivery to tumor cells.5,62 Because targets of many drugs are intracellular, synthetic receptors that promote the endocytosis of drugs might provide novel tools for drug delivery. This approach may require the development of methods to disrupt endosomal membranes to efficiently access the cellular cytoplasm and nucleus. However, synthetic cell surface receptors that irreversibly deliver proteins into degratory lysosomes might also be useful for the removal of undesirable proteins from the extracellular environment. With appropriately designed synthetic receptors, this ligand depletion approach could potentially be used to target autoreactive antibodies, tumor-promoting growth factors, or inflammatory cytokines involved in numerous diseases.
Synthetic receptors derived from N-alkyl-3β-cholesterylamines most closely mimic GPI-linked receptors and glycolipids that are anchored to the outer leaflet of the cellular plasma membrane. However, many cell surface receptors are transmembrane proteins that engage both extracellular ligands and intracellular proteins involved in signaling and endocytosis. To mimic these receptors, the development of compounds that span the plasma membrane to access intracellular proteins could provide powerful new tools for controlling myriad cellular functions including cellular proliferation and apoptosis. Previously reported steroid dimers that span lipid bilayers of liposomes and mimic aspects of cellular signaling63 provide inspiration for the design of transmembrane compounds that might function on living cells. The creation of prosthetic molecules that seamlessly access the molecular machinery of living cells represents an exciting new frontier in the fields of bioorganic chemistry and chemical biology.
Acknowledgements
I am grateful for financial support from the National Institutes of Health, the American Cancer Society, the Department of Defense Congressionally Directed Medical Research Programs, The American Chemical Society Petroleum Research Fund, and the Camille and Henry Dreyfus Foundation.References
- S. D. Conner and S. L. Schmid, Nature, 2003, 422, 37–44 CrossRef CAS.
- J. L. Goldstein, M. S. Brown, R. G. Anderson, D. W. Russell and W. J. Schneider, Annυ. Rev. Cell Biol., 1985, 1, 1–39 Search PubMed.
- M. S. Brown and J. L. Goldstein, Angew. Chem. Int. Ed. Engl., 1986, 25, 583–602 CrossRef.
- G. Rudenko, L. Henry, K. Henderson, K. Ichtchenko, M. S. Brown, J. L. Goldstein and J. Deisenhofer, Science, 2002, 298, 2353–2358 CrossRef CAS.
- G. M. Dubowchik and M. A. Walker, Pharmacol. Theor., 1999, 83, 67–123 Search PubMed.
- N. S. Chung and K. M. Wasan, Adv. Drug Delivery Rev., 2004, 56, 1315–1334 CrossRef CAS.
- E. A. Merritt, P. Kuhn, S. Sarfaty, J. L. Erbe, R. K. Holmes and W. G. Hol, J. Mol. Biol., 1998, 282, 1043–1059 CrossRef CAS.
- D. C. Smith, J. M. Lord, L. M. Roberts and L. Johannes, Semin. Cell. Dev. Biol., 2004, 15, 397–408 CrossRef CAS.
- K. Sandvig and B. van Deurs, FEBS Lett., 2002, 529, 49–53 CrossRef CAS.
- K. Sandvig and B. van Deurs, Ann. Rev. Cell Dev. Biol., 2002, 18, 1–24 CrossRef CAS.
- D. A. Brown and E. London, J. Biol. Chem., 2000, 275, 17221–17224 CrossRef CAS.
- K. Simons and E. Ikonen, Science, 2000, 290, 1721–1726 CrossRef CAS.
- F. R. Maxfield, Curr. Opin. Cell Biol., 2002, 14, 483–487 CrossRef CAS.
- M. Edidin, Annu. Rev. Biophys. Biomol. Struct., 2003, 32, 257–283 CrossRef CAS.
- D. A. Zacharias, J. D. Violin, A. C. Newton and R. Y. Tsien, Science, 2002, 296, 913–916 CrossRef CAS.
- K. Simons and D. Toomre, Nat. Rev. Mol. Cell Biol., 2000, 1, 31–39 CrossRef CAS.
- E. Ikonen, Curr. Opin. Cell Biol., 2001, 13, 470–477 CrossRef CAS.
- H. Pang, P. U. Le and I. R. Nabi, J. Cell Sci., 2004, 117, 1421–1430 Search PubMed.
- B. J. Nichols, Curr. Biol., 2003, 13, 686–690 CrossRef CAS.
- G. H. Hansen, S. M. Dalskov, C. R. Rasmussen, L. Immerdal, L. L. Niels-Christiansen and E. M. Danielsen, Biochemistry, 2005, 44, 873–882 CrossRef CAS.
- R. H. Massol, J. E. Larsen, Y. Fujinaga, W. I. Lencer and T. Kirchhausen, Mol. Biol. Cell., 2004, 15, 3631–3641 CrossRef CAS.
- R. D. Singh, V. Puri, J. T. Valiyaveettil, D. L. Marks, R. Bittman and R. E. Pagano, Mol. Biol. Cell, 2003, 14, 3254–3265 CrossRef CAS.
- S. Manes, G. del Real and A. C. Martinez, Nat. Rev. Immunol., 2003, 3, 557–568 CrossRef CAS.
- S. P. Vyas, A. Singh and V. Sihorkar, Crit. Rev. Theor. Drug Carrier Syst., 2001, 18, 1–76 Search PubMed.
- Y. Lu, E. Sega, C. P. Leamon and P. S. Low, Adv. Drug Delivery Rev., 2004, 56, 1161–1176 CrossRef CAS.
- Z. M. Qian, H. Li, H. Sun and K. Ho, Pharmacol. Rev., 2002, 54, 561–587 Search PubMed.
- S. L. Hussey, E. He and B. R. Peterson, J. Am. Chem. Soc., 2001, 123, 12712–12713 CrossRef CAS.
- S. Boonyarattanakalin, S. E. Martin, S. A. Dykstra and B. R. Peterson, J. Am. Chem. Soc., 2004, 126, 16379–16386 CrossRef CAS.
- S. L. Hussey and B. R. Peterson, J. Am. Chem. Soc., 2002, 124, 6265–6273 CrossRef CAS.
- S. E. Martin and B. R. Peterson, Bioconjugate Chem., 2003, 14, 67–74 CrossRef CAS.
- K. N. Chung, S. Roberts, C. H. Kim, M. Kirassova, J. Trepel and P. C. Elwood, Arch. Biochem. Biophys., 1995, 322, 228–234 CrossRef CAS.
- M. Hao and F. R. Maxfield, J. Biol. Chem., 2000, 275, 15279–15286 CrossRef CAS.
- F. R. Maxfield and T. E. McGraw, Nat. Rev. Mol. Cell Biol., 2004, 5, 121–132 CrossRef CAS.
- L. Cedergren, R. Andersson, B. Jansson, M. Uhlen and B. Nilsson, Protein Eng., 1993, 6, 441–448 CrossRef CAS.
- M. Hao, S. X. Lin, O. J. Karylowski, D. Wustner, T. E. McGraw and F. R. Maxfield, J. Biol. Chem., 2002, 277, 609–617 CAS.
- E. Reaven, L. Tsai and S. Azhar, J. Biol. Chem., 1996, 271, 16208–16217 CrossRef CAS.
- B. Kellam, P. A. De Bank and K. M. Shakesheff, Chem. Soc. Rev., 2003, 32, 327–337 RSC.
- M. E. Medof, S. Nagarajan and M. L. Tykocinski, FASEB J., 1996, 10, 574–586 Search PubMed.
- D. R. Premkumar, Y. Fukuoka, D. Sevlever, E. Brunschwig, T. L. Rosenberry, M. L. Tykocinski and M. E. Medof, J. Cell. Biochem., 2001, 82, 234–245 CrossRef CAS.
- M. Comiskey, C. Y. Goldstein, S. R. De Fazio, M. Mammolenti, J. A. Newmark and C. M. Warner, Hum. Immunol., 2003, 64, 999–1004 CrossRef CAS.
- G. Civenni, S. T. Test, U. Brodbeck and P. Butikofer, Blood, 1998, 91, 1784–1792 CAS.
- C. W. van den Berg, T. Cinek, M. B. Hallett, V. Horejsi and B. P. Morgan, J. Cell. Biol., 1995, 131, 669–677 CrossRef CAS.
- L. Rajendran and K. Simons, J. Cell Sci., 2005, 118, 1099–1102 Search PubMed.
- S. Nagarajan, M. Anderson, S. N. Ahmed, K. W. Sell and P. Selvaraj, J. Immunol. Methods, 1995, 184, 241–251 CrossRef CAS.
- J. de Kruif, M. Tijmensen, J. Goldsein and T. Logtenberg, Nat. Med., 2000, 6, 223–227 CrossRef CAS.
- K. Kato, C. Itoh, T. Yasukouchi and T. Nagamune, Biotechnol. Prog., 2004, 20, 897–904 CrossRef CAS.
- T. Y. Wang, R. Leventis and J. R. Silvius, J. Biol. Chem., 2005, 280, 22839–22846 CrossRef CAS.
- C. R. Bertozzi and L. L. Kiessling, Science, 2001, 291, 2357–2364 CrossRef CAS.
- D. H. Dube and C. R. Bertozzi, Curr. Opin. Chem. Biol., 2003, 7, 616–625 CrossRef CAS.
- L. K. Mahal, K. J. Yarema and C. R. Bertozzi, Science, 1997, 276, 1125–1128 CrossRef CAS.
- E. Saxon and C. R. Bertozzi, Science, 2000, 287, 2007–2010 CrossRef CAS.
- J. A. Prescher, D. H. Dube and C. R. Bertozzi, Nature, 2004, 430, 873–877 CrossRef CAS.
- J. B. Rothbard, T. C. Jessop and P. A. Wender, Adv. Drug Delivery Rev., 2005, 57, 495–504 CrossRef CAS.
- B. Gupta, T. S. Levchenko and V. P. Torchilin, Adv. Drug Delivery Rev., 2005, 57, 637–651 CrossRef CAS.
- M. Zorko and U. Langel, Adv. Drug Delivery Rev., 2005, 57, 529–545 CrossRef CAS.
- S. Futaki, Adv. Drug Delivery Rev., 2005, 57, 547–558 CrossRef CAS.
- H. Brooks, B. Lebleu and E. Vives, Adv. Drug Delivery Rev., 2005, 57, 559–577 CrossRef CAS.
- A. Fittipaldi and M. Giacca, Adv. Drug Deliv. Rev., 2005, 57, 597–608 CrossRef CAS.
- J. R. Maiolo, M. Ferrer and E. A. Ottinger, Biochim. Biophys. Acta, 2005, 1712, 161–172 CAS.
- S. M. Fuchs and R. T. Raines, Biochemistry, 2004, 43, 2438–2444 CrossRef CAS.
- M. Simonova, O. Shtanko, N. Sergeyev, R. Weissleder and A. Bogdanov, Jr., J. Gene Med., 2003, 5, 1056–1066 CrossRef CAS.
- M. Lenz, W. P. Miehe, F. Vahrenwald, G. Bruchelt, P. Schweizer and R. Girgert, Anticancer Res., 1997, 17, 1143–1146 CAS.
- P. Barton, C. A. Hunter, T. J. Potter, S. J. Webb and N. H. Williams, Angew. Chem. Int. Ed., 2002, 41, 3878–3881 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2005 |
