Structure–activity relationships, kinetics, selectivity, and mechanistic studies of synthetic hydraphile channels in bacterial and mammalian cells
W. Matthew
Leevy
a,
Seth T.
Gammon
b,
Tatiana
Levchenko
e,
David D.
Daranciang
a,
Oscar
Murillo
a,
Vladimir
Torchilin
e,
David
Piwnica-Worms
ab,
James E.
Huettner
c and
George W.
Gokel
*ad
aDepartment of Molecular Biology & Pharmacology, Washington University School of Medicine, Campus Box 8103, 660 S. Euclid Avenue, St. Louis, MO 63110, USA. E-mail: ggokel@wustl.edu; Tel: +1 314 362 9297
bDepartments of Radiology, Washington University School of Medicine, Campus Box 8103, 660 S. Euclid Avenue, St. Louis, MO 63110, USA
cDepartments of Cell Biology and Physiology, Washington University School of Medicine, Campus Box 8103, 660 S. Euclid Avenue, St. Louis, MO 63110, USA
dDepartment of Chemistry, Washington University, 1 Brookings Drive, St. Louis, MO 63130, USA
eDepartment of Pharmaceutical Sciences, Bouve College of Health Sciences, Northeastern University, Boston, MA 02115, USA
First published on 26th August 2005
Abstract
Hydraphile compounds are shown to be cytotoxic to Gram-negative and Gram-positive bacteria, yeast, and mammalian cells. Their cellular toxicity compares favorably with other synthetic ionophores and rivals that potency of natural antibiotics. The effects of structural variations on toxicity are described. The effects of these variations correlate well with previous studies of ion transport in liposomes. Whole cell patch clamping with mammalian cells confirms a channel mechanism in living cells suggesting that this family may comprise novel and flexible pharmacological agents.
Introduction
Biological membranes are complex but ubiquitous structures that comprise the boundary layer of cells and organelles. Natural bilayers are formed by contact between the non-polar hydrocarbon chains of two leaflets. The polar headgroups of phospholipids contact the aqueous cytosol or periplasm. In vital systems, the bilayer is usually asymmetric and contains numerous other species, including proteins, sterols, sphingolipids, and other lipids. Bilayer membranes provide the fundamental protective barrier between the cell and the outside environment, but they are also involved in cell respiration, nerve conduction, and many other physiological activities. Despite decades of intense research, uncertainty remains over many of their features, including the exact head group orientation and the extent of hydrocarbon interdigitation between leaflets.1Membrane-spanning proteins have garnered attention for their roles in regulating numerous cellular processes, including ion balance, cell signaling, and the uptake of organic substrates. Of particular interest have been protein channels that mediate the passage of ions through membranes. Channel proteins play the remarkable role of moving charged species through the approximate 30 Å thickness of membrane hydrocarbons. The “hydrocarbon slab” or “insulator regime” is thought to have a dielectric constant of 2–3. Nature's rigorous maintenance of cellular salt concentrations demonstrates the importance of ion transport, which must occur through the insulating bilayer. The protein ion channels that mediate ion conductance have been studied for decades, but structural details have only recently become available.2 Moreover, mechanistic details remain obscure in most cases.
Because channel proteins are so complex, chemists as early as the 1980s responded to the challenge of developing synthetic ionophores that are working models of these remarkable natural systems.3 Early contributions to the field came from the groups of Tabushi,4 Lehn,5 Fyles,6 and our own laboratory.7 More recent contributions include the peptide nanotubes of Ghadiri,8 the peptide-linked crown ethers of Voyer,9 our chloride-selective heptapeptide channels,10 and others.11 Many of the synthetic ion channel compounds have demonstrated classic open/closed channel behavior in planar bilayer voltage clamp experiments3,12,13 and have shown activity in synthetic liposomes.14–16 The peptide back-boned crown system9 is thought to form an α-helix in the membrane, which then aligns the macrocycles to form tubes through the bilayer. The cyclic D,L-amino acid nanotubes8 stack to form an extended, hydrogen-bonded tunnel that permits the passage of ions or sugars across the membrane. The hydraphiles mimic the structure of known protein channels by having an entry and exit portal connected to a centrally hydrated relay. Fig. 1 shows a schematic representation of these three ion conducting compounds or assemblies as they are thought to function in the bilayer.
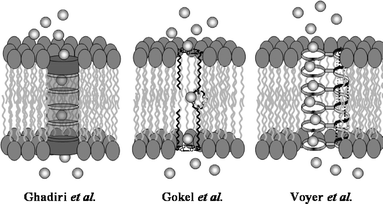 | ||
| Fig. 1 Presumed channel conformations for (left) peptide nanotubes, (center) hydraphiles, and (right) multi-crown peptide. | ||
Natural ionophores are known to exhibit toxic effects in cellular systems. Examples include valinomycin,17 the cecropins,18 gramicidin,19 and melittin.20 The synthetic ionophores described above are also biologically active. Specifically, the peptide-linked crown ethers cause red blood cell haemolysis but are inactive against E. coli,21 while the cyclic peptide nanotubes and the synthetic hydraphiles act as potent antimicrobial agents.22,23 The cyclic peptide nanotubes are quite active, killing bacteria at concentrations as low as 2.5 µM.24 The hydraphiles, studies of which are reported here, are also cytotoxic to bacteria at concentrations as low as 0.6 µM. Other synthetic channels25 presumably hold this potential, although, to our knowledge, it is currently unrealized.
The toxicity of synthetic channels is presumably the result of rapid, unregulated ion flux through the plasma membrane that disrupts cellular ion gradients causing osmotic stress and, ultimately, cell death. Two mechanisms have been proposed to describe this process.26 Previous studies using membrane potential sensitive dyes showed that hydraphiles act as ionophores in the E. coli membranes.23 The hydraphiles were designed to be modular and are available in a range of lengths and polarities. We have assayed the biological activity of 13 synthetic ionophores and report here the observed structure–activity relationships. In addition, evidence for the mechanism of action is presented for the hydraphile family of compounds.
Results and discussion
Compounds studied
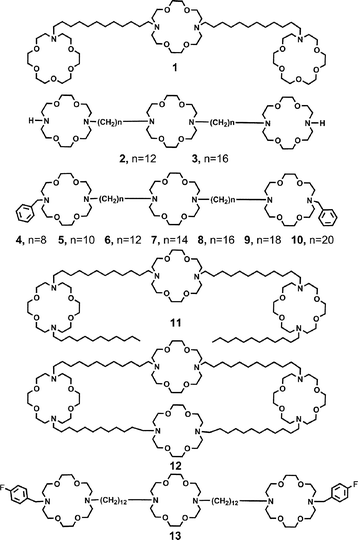
Hydraphiles are typically comprised of three diaza-18-crown-6 units separated by two 12-carbon spacers. The two distal crowns act as head groups and as portals for ion entry and egress. The central macrocycle is an ion relay27 that serves the same purpose as the “water and ion-filled capsule” recently identified in the solid state structure of the KcsA channel of Streptomyces lividans.28 The hydraphile compounds used in this study are shown as 1–13.Compounds 1–11 and 13 contain three macrocycles linked covalently. The distal macrocycles of 2–11 are 4,13-diaza-18-crown-6, but in 1 the corresponding macrocycle is aza-18-crown-6. The sidearms in structures 4–10 are benzyl and n-dodecyl in 11. Compounds 4–10 differ in spacer chain length. The shortest chain is octylene (4) and the longest is eicosylene (10). The spacer chains in tris(macrocycle)s 1, 2, 6, 11 and 13 are all dodecylene. Compound 12 differs from the other structures in this study because a fourth macrocycle is present and linked covalently to the distal macrocycles. This symmetrical structure is thought to form a “tunnel” within the bilayer and is the most active sodium ion transporter reported in this study. Finally, compound 13 is identical to 6 except that para-fluoro substituents are present on the benzyl groups.
Structural and fluorescence studies indicate that the hydraphiles arrange in the bilayer with the distal macrocycles at opposite ends of the membrane's insulator regime while the hydrocarbon chains (e.g. the dodecyl groups of 11) align with the fatty acid chains.29 Compound 1 does not possess a side chain; instead the distal macrocycles are aza-18-crown-6 residues. In this compound, the hydraphile's typical ![[double bond splayed left]](https://www.rsc.org/images/entities/char_e009.gif) N–R side chain is replaced by a single oxygen atom. Compound 1 showed no detectable sodium transport activity in 23Na-NMR experiments conducted in liposomes.14 In these same studies, compounds 2, 6, and 11 had relative rates of 28, 39, and 28 (compared to the transport activity of gramicidin (arbitrarily set to 100) determined simultaneously). Compound 12, the dodecyl chain “tunnel” structure, shows a relative rate for sodium ion transport of approximately 81 compared to the gramicidin standard.30 These sodium transport results indicate that an
N–R side chain is replaced by a single oxygen atom. Compound 1 showed no detectable sodium transport activity in 23Na-NMR experiments conducted in liposomes.14 In these same studies, compounds 2, 6, and 11 had relative rates of 28, 39, and 28 (compared to the transport activity of gramicidin (arbitrarily set to 100) determined simultaneously). Compound 12, the dodecyl chain “tunnel” structure, shows a relative rate for sodium ion transport of approximately 81 compared to the gramicidin standard.30 These sodium transport results indicate that an ![[double bond splayed left]](https://www.rsc.org/images/entities/char_e009.gif) N–H (not an ether link) is the minimum sidearm commensurate with transport activity. Other side chains and spacer chain lengths alter transport activity. These results are mirrored in the biological results described below.
N–H (not an ether link) is the minimum sidearm commensurate with transport activity. Other side chains and spacer chain lengths alter transport activity. These results are mirrored in the biological results described below.
Biological activity
Hydraphiles are cytotoxic to the Gram-negative bacterium E. coli.23 Toxicity studies (IC50 curves) showed benzyl channel (6) to be active in the 10 µM range, while the shorter C8 benzyl channel (4) was 13-fold less active. Control compounds N,N′-didodecyl-4,13-diaza-18-crown-6 and N-benzyl-4,13-diaza-18-crown-6 were both inactive to E. coli when assessed using the disk diffusion method. In addition, a compound in which the macrocyclic central relay was replaced by a biphenyl group and known27 not to transport ions in vitro, was also inactive. These controls suggest that toxicity is caused by channel formation in the membrane rather than a non-specific detergent interaction. The fluorescent dansyl channel was used in microscopy experiments to show that hydraphile compounds localized to the membrane of E. coli.Side arm effects
Compounds 1–12 were prepared and assayed for toxicity to bacteria. The toxicity results for 1–12 and their activity in synthetic liposomes were comparable. The minimum bactericidal concentration (MBC) and minimum fungicidal concentration (MFC) for each hydraphile are reported in Tables 1 and 2 as the lowest serial 2-fold dilution that killed 99.9% of a cultured bacterium or yeast, as described by the National Committee for Clinical Laboratory Standards (NCCLS).31,32 Compound 1 has a MBC of 87 µM to B. subtilis, and 175 µM to E. coli and S. cerevisiae. We consider this to be largely inactive, a result in accord with the previously noted sodium ion transport data.| Compound | |||||||
|---|---|---|---|---|---|---|---|
| 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| a Minimum bactericidal concentration (in µM). b Not determined. c Percent release of Na+ from liposomes determined by using an ISE technique.15 | |||||||
| Spacer chain length | (CH2)8 | (CH2)10 | (CH2)12 | (CH2)14 | (CH2)16 | (CH2)18 | (CH2)20 |
| E. coli | 170 | 80 | 9.4 | 2.3 | 4.6 | 75 | 160 |
| B. subtilis | 42 | 10 | 1.2 | 0.56 | 0.6 | 1 | 2 |
| S. cerevisiae | 170 | 160 | 38 | 4.6 | 2.3 | —b | —b |
| Na+ releasec | ∼2 | 23 | 84 | 100 | 95 | 39 | 14 |
Replacing the macroring oxygen of 1 by NH results in 2, which shows a significant increase in toxicity to both E. coli and B. subtilis (MBC = 22 µM and 11 µM), while the yeast remains resistant. “Benzyl channel”, 6, is formed when a benzyl group replaces the hydrogen of the N–H group of 2. The activity of 6 increases compared to 2 by about 2-fold for E. coli (MBC = 9.4 µM) and 10-fold for B. subtilis (MBC = 1.2 µM). A moderate increase in activity is also observed for S. cerevisiae (MFC = 39). The toxicity of these compounds rates well as antibiotics are generally considered effective at MBCs of 10 µM and below.33 When the distal macrocycles of the hydraphile framework are terminated by 12-carbon side chains (11), activity increases again by about two-fold to 2.1 µM for E. coli and 0.53 µM for B. subtilis. The yeast sustain an approximate 5-fold increase in activity to 8.4 µM.
Linking the dodecyl side chains through an additional macrocycle (12) leads to another two-fold enhancement in activity to ∼2 µM for E. coli, while B. subtilis remains at the 500 nM level of inhibition. We note that 12 was the most active sodium transporter studied in liposomes.30 Yeast are resistant to the action of 12 but they are susceptible to 11 (8.4 µM). Some of the difference in activity may result from differences in the environments in which the organisms were grown. S. cerevisiae are grown in YPD media that has pH = 6.5, compared to LB media (bacteria) that has pH = 7. Previous studies showed that higher acidity reduced the toxicity of 2, 6, and 12 to bacteria. Surprisingly, 11 was found to be more toxic at lower pH. The yeast are grown at a lower pH and, as noted for bacteria, 11 is the most active compound, while 2, 6, and 12, are less cytotoxic.
Hydraphile channel length effects on transport and toxicity
Membrane thickness depends on numerous factors. These include, but are not limited to, the extent of hydrocarbon chain interdigitation, the chain length of the lipid hydrocarbon chains, fatty acid unsaturation and double bond geometry, differences in head groups, and the presence or absence of such membrane thickening agents as cholesterol. In a previous study,27 we examined Na+ transport mediated by analogs of 6 that are identical except that their spacer chains vary in length. Initially, we studied sodium equilibration mediated by 4–8 in phospholipid liposomes by using the 23Na NMR method. Compounds 6 and 7 were found to be about equally active and more active than shorter or longer analogs.A more recent study expanded the range from 4–10; these compounds differ in spacer chain length from 8 to 20 methylene units. In this work, an ion selective electrode (ISE) method was used to monitor Na+ efflux in synthetic liposomes.15 The phospholipids used to prepare different vesicles were identical, including a single cis-unsaturation. The data obtained for 4–10 confirmed the trend previously observed by NMR for 4–8. In addition, hydraphile length was found to generally correlate with phospholipid fatty acid chain length, i.e., membrane thickness.15 This critical correspondence between hydraphile length and membrane thickness was explored in the biological context using E. coli, B. subtilis, and S. cerevisiae.
The benzyl channel compounds (4–10) that were studied had spacer chains of 8, 10, 12, 14, 16, 18, and 20 methylenes. Table 2 records the response of microorganisms to the presence of these compounds. The C8 benzyl channel (4) is the least active compound to the bacteria. This result mirrors those of previous studies conducted in phospholipid vesicles. A modest increase in toxicity to both E. coli and B. subtilis is observed for 5 (C10 benzyl channel), which is approximately 5 Å longer overall than is 4. Addition of two more methylenes in each spacer chain gives 6 (C12 benzyl channel) and engenders a sizable increase in activity. This further 5 Å of channel length affords a ∼9-fold increase in toxicity to E. coli and to B. subtilis. Maximal toxicity is reached for E. coli in the C14–C16 range and activity declines as length increases further (9, C18 and 10, C20 hydraphiles). These results correlate well with Na+ transport activity monitored in synthetic liposomes.34 Maximal toxicity to B. subtilis, 0.56 µM, is observed for the C14 and C16 channels (7, 8). This value of ∼0.6 µM compares with a toxic concentration for penicillin of ∼8 µM and is among the most potent examples of ionophore-mediated toxicity reported, natural or synthetic.
Kinetics of toxicity to E. coli determined by using bioluminescent bacteria
The potent toxicity of the hydraphiles to both E. coli and B. subtilis is apparent from the final MBC values. How rapidly the bacteria are killed is a question that could be addressed directly by use of bioluminescent bacteria. E. coli bacteria expressing the lux operon genes of Photorhabdus luminescens on a pT7-3 plasmid were used for the kinetic study. These bioluminescent bacteria emit light during log phase growth and the disappearance of light is a quantitative measure of cell death.35 The experiments reported here monitored the loss of photon emission from the luminescent E. coli after treatment with hydraphile at the concentration determined for the MBC. The studies were performed in 96-well plates and photon output was read using a CCD camera (see Experimental section). Fig. 2 shows the decay of light from the bacteria as a percentage of the light emitted from untreated control wells.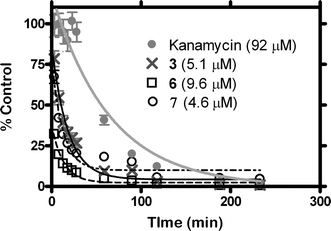 | ||
| Fig. 2 Kinetics of killing bioluminescent E. coli with 3, 6, and 7 at the indicated concentrations. Also shown are data for kanamycin. | ||
These data highlight the potency of hydraphiles. When tested at their MBCs, 6, 7, and 3 killed half of the E. coli population in 8.5, 9.1, and 12.5 minutes. This compares with a halftime of the known antibiotic kanamycin (MBC = 1.3 µM) of 44.8 minutes at 10–20 times the concentration of hydraphile. Although less active than the hydraphiles, kanamycin shows high selectivity for bacteria over mammalian cells, while the hydraphiles do not (see below).
Hydraphile aggregation
Some amphiphilic molecules are known to exhibit toxicity through membrane disruption effects caused by the formation of micelles. A study of hydraphile aggregation in buffer was conducted to determine their critical micelle concentrations (CMCs). Compound 6 was tested at concentrations as high as 38 µM in buffer (0.5 mM HEPES, 67 mM KCl), and no aggregation was detected by laser light scattering analysis (see Experimental section). The benzyl channel derivatives typically exhibit cytotoxicity to bacteria in the 0.5–10 µM range. Since this range is significantly below 38 µM, we conclude that aggregation of hydraphiles in solution does not influence their toxicity.Mammalian cell cytotoxicity
Toxicity studies were performed on Human Embryonic Kidney (HEK 293) and colonic carcinoma (CaCo2) cells to ascertain the inherent selectivity of hydraphiles between mammalian and bacterial cells, and also to assess anti-tumor efficacy. HEK 293 cells were incubated with 6 and 7 (C12 and C14 benzyl channels) for 24 h and assayed for their ability to reduce 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium salt (MTS assay).36 The results using HEK 293 cells are shown graphically in Fig. 3. Hydraphiles 3, 5, and 6 have LD50 values in the range of 2 µM. The effectiveness of hydraphile channel compounds across all types of cells is apparent in these data, as both bacteria and mammalian cell types are killed in the same concentration range.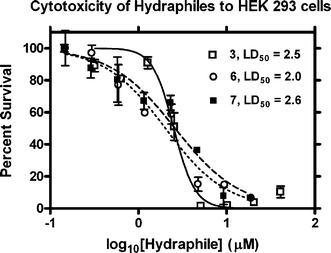 | ||
| Fig. 3 Toxicity of hydraphiles 3, 6, and 7 to HEK 293 cells. | ||
This MTS assay was further used to assess the antitumor activity of compounds 4, 6, and 11 against CaCo2 cells. Compounds 6 and 11 showed LD50 values of ∼12 µM while the C8 benzyl hydraphile, 4, was inactive at concentrations as high as 20 µM (Fig. 4). These CaCo2 cancer cells are about 6-fold more resistant to 6 than are HEK cells. A possible explanation is that the apoptosis machinery normally present in non-cancer cells such as HEK 293 is suppressed.
![Cytotoxicity of hydraphiles to CaCo2 cells. [Hydraphile] is micromolar.](/image/article/2005/OB/b508157b/b508157b-f4.gif) | ||
| Fig. 4 Cytotoxicity of hydraphiles to CaCo2 cells. [Hydraphile] is micromolar. | ||
The use of electrophysiology to probe mechanism
The whole cell patch clamping technique is widely used and accepted by biologists to study ion channel behavior. We have used this method to show that indeed hydraphiles are acting as ionophores in the bilayers of living organisms, and generate levels of current on a par with native protein ion channels present in the bilayer. Whole cell patch clamping experiments were undertaken to evaluate the membrane channel activity of hydraphiles in a biological setting and to better understand their behavior in the bilayer of a living cell.37 HEK 293 cells were held in whole cell mode at −20 mV (with respect to the outside of the cell) giving the baseline current represented as the solid line shown in the left panel of Fig. 5. The points during each phase of the experiment represent 10 ms voltage steps from −100 to +60 mM, and appear as a continuous line during saline rinse.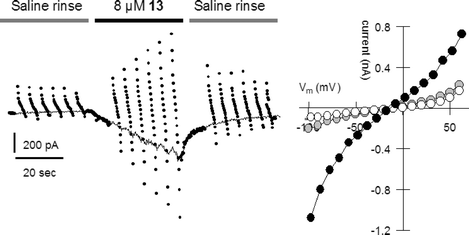 | ||
| Fig. 5 Cellular electrical response during a brief application of 13 to an HEK 293 cell (black bar) under whole cell patch clamp conditions. | ||
The left panel of Fig. 5 shows the membrane current of HEK 293 cells during rinse with saline solution, followed by a brief application of p-fluorobenzyl hydraphile 13. This compound is identical to 6 (illustrated) except that a fluorine atom is present in the para- or 4-position of the benzyl group. After application of 13, the cell is rinsed with saline solution. Compound 13 was tested because it had previously shown long open times during planar bilayer conductance experiments.38
The right panel of Fig. 5 shows the current–voltage (I–V) plot of the cell during its first rinse with saline solution (open circles), during treatment with 13 (black circles), and during extended rinsing (grey circles). Membrane conductance increases from about 1 nS to 7 nS upon treatment with 4-fluorobenzyl compound 13. The cell exhibited a quick return to homeostasis after hydraphile application was stopped. Even after prolonged rinsing, a residual conductance of 1.8 nS persisted, possibly indicating stable inclusion of the compound in the cellular bilayer. On average, application of the compound caused a 3.7 ± 1.2 fold (n = 4) increase in membrane conductance. Based on these results, the use of hydraphiles at subtoxic concentrations may hold utility in the recently posited pharmaceutical approach known as channel replacement therapy.39
The HEK 293 cells are bathed in Tyrode's solution, which contains 150 mM NaCl as the highest concentration salt. The cells are clamped at an inside potential of −20 mV with respect to the outside. Thus, positively charged Na+ ions pass into the cell through a hydraphile, while negatively charged particles depart. The concentrations of Na+ and Cl− are low within the cell but the K+ concentration is high. Glucuronate, a large organic anion, is also present. The latter presumably cannot pass through the hydraphile channel. We therefore reason that Na+ is the primary ion driving the sizable currents noted at negative holding potentials. When the cell is clamped at positive potentials, we observe sizable currents equal to those just noted. These currents may be carried by K+ leaving the cell, and/or by Cl− entering the cell.
Conclusion
The present study demonstrates the potent cytotoxicity of hydraphile compounds to Gram-negative and Gram-positive bacteria, yeast, and mammalian cells. These compounds compare favorably with other synthetic ionophores in toxicity, and rival those developed by nature over millions of years. Flexible synthetic access to these compounds allows structural components such as length to be optimized to maximize toxicity. Further, sidearms may be changed completely or adjusted to vary steric bulk, hydrophobicity, electron richness, etc. The structural dependence of toxicity corresponds well with our previous ion transport experiments utilizing liposomes. In addition, whole cell patch clamping with mammalian cells confirms a channel mechanism in these living systems. The hydraphile class of compounds provides a functional model to study channel mediated ion transport in a biological setting, and furthermore offers a strong architecture for the pursuit of novel and flexible pharmacological agents.Experimental
General
1H-NMR were recorded at 300 MHz in CDCl3 solvents and are reported in ppm (δ) downfield from internal (CH3)4Si. 13C-NMR were recorded at 75 MHz in CDCl3 unless otherwise stated. Infrared spectra were recorded on a Perkin-Elmer 1710 Fourier transform infrared spectrophotometer and were calibrated against the 1601 cm−1 band of polystyrene. Melting points were determined on a Thomas Hoover apparatus in open capillaries and are uncorrected. Thin layer chromatographic (TLC) analyses were performed on aluminium oxide 60 F-254 neutral (Type E) with a 0.2 mm layer thickness or on silica gel 60 F-254 with a 0.2 mm layer thickness. All reactions were conducted under dry N2 unless otherwise stated. All reagents were the best (non-LC) grade commercially available and were distilled, recrystallized, or used without further purification, as appropriate. Combustion analyses were performed by Atlantic Microlab, Inc., Atlanta, GA, and are reported as percents.N-4-Fluorobenzyl-4,13-diaza-18-crown-6. Diaza-18-crown-6 (3.55 g, 13.5 mmol), 4-fluorobenzyl bromide (2.20 g, 11.6 mmol), Na2CO3 (28.8 g, 272 mmol), KI (0.32 g, 1.9 mmol), and CH3CH2CH2CN (135 mL) were heated under reflux (72 h) in a 250 mL round bottomed flask. The reaction mixture was evaporated and the residue dissolved in CH2Cl2 (100 mL), washed with H2O (3 × 50 mL), dried (MgSO4), and evaporated. Chromatography over Al2O3 (2% 2-PrOH in CH2Cl2) gave the title compound (1.14 g, 41%) as a white solid (mp 83–84 °C). NMR: 2.8–2.9 (m, 8H); 3.6–3.7 (m, 19H); 6.95–7.05 (pseudo-T, 2H); 7.3–7.4 (pseudo-T, 2H).
7,16-Bis-{12-[16-(4-Fluorobenzyl)-1,4,10,13-tetraoxa-7,16-diazacyclooctadec-7-yl]-dodecyl}-1,4,10,13-tetraoxa-7,16-diazacyclooctadecane, 13. N-4-Fluorobenzyldiaza-18-crown-6 (0.8 g, 2.2 mmol), N,N′-di-(n-12-bromododecyl)-4,13-diaza-18-crown-6 (0.82 g, 1.08 mmol), Na2CO3 (2.3 g, 21.7 mmol), KI (14 mg), and CH3CH2CH2CN (14 mL) were heated at reflux for 25 h. The reaction mixture was evaporated to a thick oil (1.98 g). Column chromatography (Al2O3, 10–15% 2-propanol in hexanes) gave 13 (0.41 g, 28%) as a waxy solid. NMR: 1.2–1.6 (m, 40H), 2.5–2.6 (broad S, 8H), 2.7–3.0 (m, 24H), 3.5–3.7 (broad s, 52H), 7.0, 7.3–7.4 (m, m, 8 H). Calculated for C74H132F2N6O12, High resolution mass spectrometry, exact mass: 1334.99. Found, (M + 1)+m/z 1335.996.
Electrophysiology
The methods used were those reported previously.37Microbial cytotoxicity
The minimum inhibitory concentration (MIC) for each hydraphile is reported as the lowest serial 2-fold dilution that prevented bacterial growth as outlined by the National Committee for Clinical Laboratory Standards (NCCLS).31,32E. coli DH5α cells with a pBluescript plasmid having AMP resistance were tested using the standard inoculum size of 5 × 105 CFU mL−1. Cells were grown at 37 °C in 2 mL of Luria Bertani (LB) Miller media (10 g L−1 peptone, 5 g L−1 yeast extract, 10 g L−1 NaCl, 100 µg mL−1 ampicillin) that were 2-fold serially diluted with hydraphile test compound. Bacillus subtilis (JH642 WT) cells were tested in similar fashion using Luria broth (no AMP) at the same inoculum size, and grown at 30 °C. The MIC was taken as the lowest hydraphile concentration that inhibited growth after 24 h as judged by visual turbidity. Each compound was assayed three times at every reported concentration to each bacterium, using an independent dilution of compound for each experiment. The minimum bactericidal concentration (MBC) was determined as the lowest macrodilution that killed >99.9% of bacteria in the culture. This was determined by plating aliquots of the test suspensions onto Petri dishes and counting the number of colony forming units after overnight growth.Saccharomyces cerevisiae MIC/MFC experiments were conducted in similar fashion. Cells were grown in YPD media (10 g L−1 yeast extract, 20 g L−1 peptone, 20 g L−1 dextrose) at 30 °C. An inoculum of 5 × 103 cells was used for MIC studies, and allowed to grow for 48 hours to achieve turbidity.
CaCo2 Cytotoxicity
CaCo2 cells were seeded in 96-well tissue culture microtiter plates at a density of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well. After 24 h, the culture medium was removed and the cells were treated with 4, 6, and 11 in serum-free medium. After 24 h, CellTiter 96 Aqueous One solution (Promega) was added to each well, and the plates re-incubated for 4 h. This assay is based on the bioreduction of MTS tetrazolium compound (Owen's reagent) into a colored soluble formazan product. The viability of cells was measured using a plate-reader (Multiscan MCC/340, Fisher Scientific) at 490 nm. Relative viability was calculated from reference cells treated with medium alone as a control.
000 cells per well. After 24 h, the culture medium was removed and the cells were treated with 4, 6, and 11 in serum-free medium. After 24 h, CellTiter 96 Aqueous One solution (Promega) was added to each well, and the plates re-incubated for 4 h. This assay is based on the bioreduction of MTS tetrazolium compound (Owen's reagent) into a colored soluble formazan product. The viability of cells was measured using a plate-reader (Multiscan MCC/340, Fisher Scientific) at 490 nm. Relative viability was calculated from reference cells treated with medium alone as a control.
HEK 293 cytotoxicity assay
HEK 293 cells were regularly cultured in a 5% CO2 100% humidity tissue culture incubator. Cells were split regularly into DMEM (Invitrogen) supplemented with 10% heat inactivated fetal bovine serum (ΔFBS), 1% Gln, 0.1% penicillin/streptomycin–0.1% fungizone.After being freshly split, cells were counted on a hemacytometer and plated at a density of 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well in a 96-well plate and grown for 24 hours. Ethanol stocks of each compound were diluted 1 : 100 into DMEM supplemented with 10%
ΔFBS and 1% Gln in triplicate. They were then serially diluted ½ into DMEM supplemented with 10%
ΔFBS, 1% Gln, and 1% ethanol. The original media was then removed from the cells and replaced with media containing the desired concentration of compound. As a positive control for growth, three wells containing cells were treated with DMEM supplemented with 10%
ΔFBS, 1% Gln, and 1% ethanol. As a negative control, three wells without cells were treated with DMEM supplemented with 10%
ΔFBS, 1% Gln, and 1% ethanol. The remaining empty wells on the plate were filled with 100 µL of phosphate buffered saline to minimize evaporation. After another 24 hours of culture, 20 µL of Cell Titer 96 Aqueous One (Promega) was added to each well and then developed in the tissue culture hood between 1 and 2 hours as per the manufacturer's protocol. The absorbance was measured at 490 nm and at 630 nm to correct for nonspecific absorbance. The data were plotted as OD490–OD630 and fit to a sigmoidal dose response curve using GraphPad 4 software.
000 cells per well in a 96-well plate and grown for 24 hours. Ethanol stocks of each compound were diluted 1 : 100 into DMEM supplemented with 10%
ΔFBS and 1% Gln in triplicate. They were then serially diluted ½ into DMEM supplemented with 10%
ΔFBS, 1% Gln, and 1% ethanol. The original media was then removed from the cells and replaced with media containing the desired concentration of compound. As a positive control for growth, three wells containing cells were treated with DMEM supplemented with 10%
ΔFBS, 1% Gln, and 1% ethanol. As a negative control, three wells without cells were treated with DMEM supplemented with 10%
ΔFBS, 1% Gln, and 1% ethanol. The remaining empty wells on the plate were filled with 100 µL of phosphate buffered saline to minimize evaporation. After another 24 hours of culture, 20 µL of Cell Titer 96 Aqueous One (Promega) was added to each well and then developed in the tissue culture hood between 1 and 2 hours as per the manufacturer's protocol. The absorbance was measured at 490 nm and at 630 nm to correct for nonspecific absorbance. The data were plotted as OD490–OD630 and fit to a sigmoidal dose response curve using GraphPad 4 software.
Kinetics of hydraphile cytotoxicity
TOP10 E. coli were transformed with the pT7-3 plasmid containing the lux operon from Photorhabdus luminescens. A single colony of the transformed E. coli was picked from a freshly streaked LB-Amp (100 µg mL−1) plate. The culture was grown in LB-Amp (100 µg mL−1) at 37 °C until the optical density was 0.5. The culture was then diluted to an OD = 0.1 and 50 µL was added to wells in a black-walled 96-well plate.To study the dose response of hydraphile toxicity, each hydraphile was dissolved in neat ethanol to a concentration of 10 mg mL−1 for 3 and 2.5 mg mL−1 for 6 and 7. The hydraphile ethanol stocks were then diluted 1:50 into LB-Amp (100 µg mL−1) in triplicate. As a vehicle control, ethanol was also diluted into LB-Amp (100 µg mL−1) in triplicate. The initial 1 : 50 dilution of hydraphile into LB was then further serially diluted 1 : 2 into LB-Amp (100 µg mL−1) and 2% ethanol (to maintain a constant concentration of vehicle). Finally, the procedure was repeated for kanamycin (20 mg mL−1 dissolved milli-Q water) except that the vehicle control was a 1 : 50 dilution of milli-Q water into LB-Amp (100 µg mL−1), and the media for the serial dilution was LB-Amp (100 µg mL−1) 2% milli-Q water.
The diluted compound (50 µL) was then added using a multichannel pipette to the appropriate wells containing the transformed E. coli. The result was that each well contained: 100 µL of lux operon transformed E. coli at OD = 0.05 in LB-Amp (100 µg mL−1), 1% ethanol, and the specified concentration of either hydraphile or Kanamycin.
The plates were then imaged for photon emission from the transformed E. coli by using a commercial IVIS 100 (Xenogen) CCD camera and computer system (settings: no filter, binning 8, f-stop = 1, exposure time = 30 s at the indicated time points). For the first 30 min, the plates remained in the IVIS at 37 °C, and then were transferred to an incubator in which the bacteria were grown at 37 °C with gentle shaking (∼210 rpm) between time points.
Data were analyzed by normalizing to the pre-drug photon flux from each well and then expressed as a percent of the vehicle treated wells at any given time point [eqn. (1)]. The standard error of the mean was propagated through eqn (1).
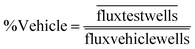 | (1) |
Acknowledgements
We thank the NIH for grants GM-36262 to GWG, NS-30888 to JEH, and P50 CA94056 to DPW that supported this work and for a Training Grant Fellowship T32-GM04892 to WML.References
- J. F. Nagle and S. Tristam-Nagle, Curr. Opin. Struct. Biol., 2000, 10, 474–480 CrossRef CAS.
- Y. Jiang, A. Lee, J. Chen, M. Cadene, B. T. Chait and R. MacKinnon, Nature, 2002, 417, 523–526 CrossRef CAS.
- G. W. Gokel and A. Mukhopadhyay, Chem. Soc. Rev., 2001, 30, 274–286 RSC.
- I. Tabushi, Y. Kuroda and K. Yokota, Tetrahedron Lett., 1982, 4601–4604 CrossRef CAS.
- L. Jullien and J.-M. Lehn, Tetrahedron Lett., 1988, 3803–3806 CrossRef CAS.
- V. E. Carmichael, P. J. Dutton, T. M. Fyles, T. D. James, J. A. Swan and M. Zojaji, J. Am. Chem. Soc., 1989, 111, 767–769 CrossRef CAS.
- A. Nakano, Q. Xie, J. V. Mallen, L. Echegoyen and G. W. Gokel, J. Am. Chem. Soc., 1990, 112, 1287–9 CrossRef CAS.
- J. D. Hartgerink, J. R. Granja, R. A. Milligan and M. R. Ghadiri, J. Am. Chem. Soc., 1996, 118, 43–50 CrossRef CAS.
- N. Voyer, L. Potvin and E. Rousseau, J. Chem. Soc., Perkin Trans. 2, 1997, 8, 1469–1471 RSC.
- P. H. Schlesinger, R. Ferdani, J. Liu, J. Pajewska, R. Pajewski, M. Saito, H. Shabany and G. W. Gokel, J. Am. Chem. Soc., 2002, 124, 1848–1849 CrossRef CAS.
- G. W. Gokel, P. H. Schlesinger, N. K. Djedovic, R. Ferdani, E. C. Harder, J. Hu, W. M. Leevy, J. Pajewska, R. Pajewski and M. E. Weber, Bioorg. Med. Chem, 2004, 12, 1291–1304 CrossRef CAS.
- E. Abel, E. S. Meadows, I. Suzuki, T. Jin and G. W. Gokel, Chem. Commun., 1997, 1145–1146 RSC.
- M. R. Ghadiri, J. R. Granja and L. K. Buehler, Nature, 1994, 369, 301–304 CrossRef CAS.
- O. Murillo, S. Watanabe, A. Nakano and G. W. Gokel, J. Am. Chem. Soc., 1995, 117, 7665–7669 CrossRef CAS.
- M. E. Weber, P. H. Schlesinger and G. W. Gokel, J. Am. Chem. Soc., 2005, 126, 636–642 CrossRef.
- N. Voyer and M. Robitaille, J. Am. Chem. Soc., 1995, 117, 6599–6600 CrossRef CAS.
- M. A. Andersson, R. Mikkola, R. M. Kroppenstedt, F. A. Rainey, J. Peltola, J. Helin, K. Sivonen and M. S. Salkinoja-Salonen, Appl. Environ. Microbiol, 1998, 64, 4767–73 CAS; K. A. Mereish, R. Solow, Y. Singh and R. Bhatnager, Med. Sci. Res., 1989, 17, 869–871 Search PubMed.
- V. C. A. Matanic and V. Castilla, Int. J. Antimicrob. Agents, 2004, 23, 382–389 CrossRef.
- H. W. Huang, Novartis Found. Symposium: Gramicidin and Related Ion Channel-Forming Peptides, 1999, 225, 188–206 Search PubMed.
- L. Béven and H. Wròblewski, Res. Microbiol., 1997, 148, 163–175 CrossRef CAS.
- Y. R. Vandenburg, B. D. Smith, E. Biron and N. Voyer, Chem. Commun., 2002, 1694–1695 RSC.
- S. A. Fernandez-Lopez, H.-S. Kim, E. C. Choi, M. Delgado, J. R. Granja, A. Khasanov, K. Kraehenbuehl, G. Long, D. A. Weinberger, K. M. Wilcoxen and M. R. Ghadiri, Nature, 2001, 412, 452–455 CrossRef.
- W. M. Leevy, G. M. Donato, R. Ferdani, W. E. Goldman, P. H. Schlesinger and G. W. Gokel, J. Am. Chem. Soc, 2002, 124, 9022–9023 CrossRef CAS; W. M. Leevy, M. R. Gokel, G. B. Hughes-Strange, P. H. Schlesinger and G. W. Gokel, New J. Chem., 2005, 1, 205–209 RSC.
- This value is based on a molecular weight of 1214 for KQRWLWLW at 3 µg mL−1 for B. subtilis.
- G. W. Gokel and O. Murillo, Acc. Chem. Res., 1996, 29, 425–32 CrossRef CAS.
- Y. Shai, Curr. Pharm. Des., 2002, 8, 715–725 Search PubMed.
- (a) C. L. Murray and G. W. Gokel, Chem. Commun., 1998, 2477–2478 RSC; (b) C. L. Murray, H. Shabany and G. W. Gokel, Chem. Commun., 2000, 2371–2372 RSC.
- D. A. Doyle, J. M. Cabral, R. A. Pfuetzner, A. Kuo, J. M. Gulbis, S. L. Cohen, B. T. Chait and R. MacKinnon, Science, 1998, 280, 69–77 CrossRef CAS.
- E. Abel, G. E. M. Maguire, O. Murillo, I. Suzuki, S. L. De Wall and G. W. Gokel, J. Am. Chem. Soc, 1999, 121, 9043–9052 CrossRef CAS.
- H. Shabany and G. W. Gokel, Chem. Commun., 2000, 2373–2374 RSC.
- National Committee for Clinical Laboratory Standards Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that grow Aerobically, 5th edn, M7–A5, NCCLS, Wayne, Pennsylvania, 2000 Search PubMed.
- National Committee for Clinical Laboratory Standards Reference Method for Broth Dilution Antifungal Susceptibility Testing of Yeast, M27–A2, NCCLS, Wayne, Pennsylvania, 2002 Search PubMed.
- H. Hanberger, L. E. Nilsson, B. Claesson, A. Kärnell, P. Larsson, M. Rylander, E. Svensson, M. Sörberg and L. Sörén, J. Antimicrob. Chemother., 1999, 44, 611–619 CrossRef CAS.
- W. M. Leevy, M. E. Weber, P. H. Schlesinger and G. W. Gokel, Chem. Commun., 2005, 89–91 RSC.
- V. Salisbury, A. Pfoestl, H. Wiesinger-Mayr, R. Lewis, K. E. Bowker and A. P. MacGowen, J. Antimicrob. Chemother., 1999, 43, 829–832 CrossRef CAS.
- G. Malich, B. Markovic and C. Winder, Toxicology, 1997, 124, 179–192 CrossRef CAS.
- W. M. Leevy, J. E. Huettner, R. P. Pajewski, P. H. Schlesinger and G. W. Gokel, J. Am. Chem. Soc., 2004, 126, 15747–15753 CrossRef CAS.
- C. L. Murray, E. S. Meadows, O. Murillo and G. W. Gokel, J. Am. Chem. Soc., 1997, 119, 7887–7888 CrossRef CAS.
- G. A. Cook, O. Prakash, K. Zhang, L. P. Shank, W. A. Takeguchi, A. Robbins, Y.-X. Gong, T. Iwamoto, B. D. Schultz and J. M. Tomich, Biophys. J., 2004, 86, 1424–1435 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2005 |