DOI:
10.1039/B504759E
(Paper)
Org. Biomol. Chem., 2005,
3, 2534-2542
Synthesis and evaluation of a new non-fluorescent quencher in fluorogenic oligonucleotide probes for real-time PCR
Received 6th April 2005, Accepted 11th May 2005
First published on 7th July 2005
Abstract
A non-fluorescent quencher, based on the diaminoanthraquinone Disperse Blue 3, has been incorporated into oligonucleotides at the 5′-end, the 3′-end and internally as a thymidine derivative. Fluorimetry and fluorogenic real-time PCR experiments demonstrate that the quencher is effective with a wide range of fluorescent dyes. The anthraquinone moiety increases the melting temperature of DNA duplexes, thus allowing shorter, more discriminatory probes to be used. The quencher has been used in Scorpion primers and TaqMan
® probes for human DNA sequence recognition and mutation detection.
Introduction
Fluorescence methods are used widely to detect specific nucleic acid sequences in homogeneous closed-tube genetic assays. Typically an oligonucleotide probe is doubly labelled with a fluorophore and a proximal quencher molecule and hybridisation of the probe to the target nucleic acid leads to signal generation
via a conformational change in the probe. Examples are Scorpion primers™
1–4 and molecular beacons.
5,6 TaqMan
® probes have a different mode of action that relies upon enzymatic cleavage to separate the fluorophore and quencher.
7,8 The quenching mechanism in Scorpions and molecular beacons is primarily collisional in nature, whereas in TaqMan
® probes, “through-space” Förster quenching predominates. Many conventional dye/quencher pairs used in fluorogenic primers and probes suffer from drawbacks such as intrinsic fluorescence of the quencher and poor spectral overlap between the fluorescent dye and the quencher molecule. Both of these have an adverse effect on the fluorescent signal-to-noise ratio. Indeed, the non-fluorescent diazo-dyes DABCYL
6 and methyl red,
9 which are frequently used in real-time PCR probes, do not efficiently quench the fluorescence of several commonly used “long-wavelength” fluorophores such as the cyanine dye, Cy5. These dyes are widely used in fluorescent probes and are particularly important in DNA microarrays. A new range of quenchers have been developed based on azo dyes (BHQ 1, 2 and 3, Biosearch Technologies) for use with commonly used fluorescent dyes.
Recently we described a new fluorescence quencher based on 1,4-diaminoanthraquinone.
10 It is a non-fluorescent molecule, thus eliminating any possibility of background signals which can arise when fluorophores are used as quenchers. Importantly, it has a broad absorption in the 550–700 nm region of the visible spectrum and is thus able to quench a wide range of fluorophores, especially those that emit at longer wavelengths. Here, we report the synthesis of three versions of this quencher suitable for incorporation into oligonucleotide probes at either the 3′-end, 5′-end or internally in place of a thymidine. In each case the degree of fluorescence quenching has been determined. The use of the new dark quencher in TaqMan
® and Scorpion oligonucleotide probes in real-time PCR has been demonstrated and its effect on duplex stability has also been studied by UV thermal melting experiments.
Results and discussion
Dark quencher phosphoramidite: synthesis and evaluation
Commercially available 1-((2-hydroxyethyl)amino)-4-(methylamino)anthraquinone (Disperse Blue 3) was purchased from Aldrich as a crude mixture (20% purity). It was purified by silica gel column chromatography, and converted to phosphoramidite
1 for 5′-labelling of oligonucleotides during solid-phase oligonucleotide synthesis (
Scheme 1).
11,12 |
| | Scheme 1 Synthesis of Disperse Blue phosphoramidite for 5′-oligonucleotide labelling.
Reagents and conditions: i, 2-cyanoethyl-
N,
N-diisopropyl chlorophosphine (1.1 eq.), DIPEA (4 eq.), CH
2Cl
2, rt, 1 h, 64%.
| |
This monomer was used to synthesise oligonucleotide ODN 01 (
Table 1) labelled with a 5′-Disperse Blue quencher. This was compared with a 5′-methyl red labelled oligonucleotide of the same sequence (ODN 07) which was synthesised using the monomer shown in
Fig. 1.
9 Five complementary fluorescent oligonucleotides (ODN 02–06) were also synthesised, each labelled with a different fluorophore at the 3′-end. The following fluorescent dye labels were chosen to span a broad range of the visible fluorescence emission spectrum: FAM (514 nm), Cy3 (564 nm), TAMRA (584 nm), Cy5 (666 nm) and Cy5.5 (704 nm).
Fig. 2 shows the basis of the fluorescence quenching assay. When the two oligonucleotides are annealed, the fluorophore and quencher are held in close proximity causing the duplex to be non-fluorescent. Heating causes the strands to dissociate, separating the fluorophore from the quencher, and the change in fluorescence is measured. The efficiency of the quenching is expressed as a ratio of the fluorescence of the single stranded oligonucleotide (at 75 °C) relative to that of the duplex (at 25 °C). The results (
Fig. 3) were normalised to the values obtained for Disperse Blue to allow a direct comparison between each fluorophore-quencher pair. Methyl red was marginally more efficient than Disperse Blue as a quencher for FAM (
λem 514 nm), but much less efficient in quenching the other four fluorescent dyes TAMRA (
λem 584 nm), Cy3 (
λem 564 nm), Cy5 (
λem 666 nm) and Cy5.5 (
λem 704 nm). The improved quenching by Disperse Blue is likely to be a result of the relatively good overlap between its absorption spectrum and the emission spectra of the individual fluorescent dyes (
Fig. 4). In contrast, the absorption spectrum of methyl red does not overlap with the emission spectra of the longer wavelength dyes used in fluorogenic assays and it is noteworthy that methyl red does not quench Cy5 or Cy5.5 efficiently, even though the two molecules are in close proximity and are presumably able to partake in collisional quenching.
Table 1 Oligonucleotides used in this study
| Oligonucleotide | Sequence |
|---|
| D5 = 5′ Disperse Blue quencher;
D3 = 3′ Disperse Blue quencher;
MeRed = methyl red quencher;
FAM = fluorescein;
TAMRA = tetramethylrhodamine;
JOE = 4′-5′-dichloro-2′,7′-dimethoxyfluorescein;
ROX = rhodamine;
DU = Disperse Blue quencher 2′-deoxyuridine;
H = hexaethyleneglycol.
|
|---|
| ODN 01 | D5GAAAAAACTTGGATCC
|
| ODN 02 | GGATCCAAGTTTTTTC
TAMRA |
| ODN 03 | GGATCCAAGTTTTTTC
Cy3T
|
| ODN 04 | GGATCCAAGTTTTTTC
FAM |
| ODN 05 | GGATCCAAGTTTTTTC
Cy5T
|
| ODN 06 | GGATCCAAGTTTTTTC
Cy5.5T
|
| ODN 07 | MeRedGAAAAAACTTGGATCC
|
| ODN 08 | FAMATGCCCTCCCCCATGCCATCCTGCGTT
D3 |
| ODN 09 | FAMATGCCCTCCCCCATGCCATCCTGCGTT
MeRed |
| ODN 10 | TAMRAATGCCCTCCCCCATGCCATCCTGCGTT
D3 |
| ODN 11 | TAMRAATGCCCTCCCCCATGCCATCCTGCGTT
MeRed |
| ODN 12 | JOEATGCCCTCCCCCATGCCATCCTGCGTT
D3 |
| ODN 13 | JOEATGCCCTCCCCCATGCCATCCTGCGTT
MeRed |
| ODN 14 | ROXATGCCCTCCCCCATGCCATCCTGCGTT
D3 |
| ODN 15 | ROXATGCCCTCCCCCATGCCATCCTGCGTT
MeRed |
| ODN 16 | TCCAAGTTTTT
DUCTA
|
| ODN 17 | TCCAAG
DUTTTTTCTA
|
| ODN 18 | TCCAAGTTTTTTC
DUA
|
| ODN 19 | TCCAAGTTTTTTCTA |
| ODN 20 | FAMTAGAAAAAACTTGGA
|
| ODN 21 | TAGAAAAAACTTGGA |
| ODN 22 | ATTAGAAAAAACTTGGA |
| ODN 23 | FAMCCCGCGCCTTTCCTCCACTGTTGCGCGCGGG
MeRedHATGGTGTGTCTTGGGATTCA
|
| ODN 24 | FAMCCCGCGCCTTTCCTCCACTGTTGCGCGCGGG
DUHATGGTGTGTCTTGGGATTCA
|
| ODN 25 | CAGTGGAGGAAAG
MeRed |
| ODN 26 | CAGTGGAGGAAAG
D3 |
| ODN 27 | FAMCTTTCCTCCACTGTTGC
HATGGTGTGTCTTGGGATTCA
|
| ODN 28 | AGGTGCGATTCTATGAGACG
MeRedT
|
| ODN 29 | AGGTGCGATTCTATGAGACG
D3 |
| ODN 30 | FAMCGTCTCATAGAATCGCACCT
HAGATTGCCAAGGACGACATC
|
| ODN 31 | Cy3CGTCTCATAGAATCGCACCT
HAGATTGCCAAGGACGACATC
|
| ODN 32 | Cy5CGTCTCATAGAATCGCACCT
HAGATTGCCAAGGACGACATC
|
 |
| | Fig. 1 Methyl red phosphoramidite monomer. | |
 |
| | Fig. 2 Assay to determine the relative efficiency of quenchers with a range of fluorophores. When the fluorophore and quencher probes are hybridised, the fluorescence is quenched. Fluorescence is observed when the duplex is denatured by heating. | |
 |
| | Fig. 3 A comparison of the fluorescent signal-to-noise ratio of Disperse Blue (blue) and methyl red (red) quenchers for five different fluorescent dyes using the assay in
Fig. 2.
| |
 |
| | Fig. 4 The visible absorption spectra of the Disperse Blue (blue) and methyl red (red) quenchers. | |
3′-Labelling of oligonucleotides with Disperse Blue
In order to synthesise oligonucleotides labelled at the 3′-end with Disperse Blue, a suitably functionalised CPG (controlled pore glass) was prepared by the method in
Scheme 2. The 2′-deoxyribose derivative
4 (α-anomer) was prepared by a method previously reported in our laboratory
9 and this provided the backbone for attachment of the quencher moiety. In order to couple Disperse Blue to backbone
4, it was necessary to convert the primary alcohol to an amine. This was carried out in high yield
via a Mitsunobu reaction, followed by hydrazine cleavage of the phthalimide protecting group. The primary amino group of compound
2 was then treated with succinic anhydride to produce the Disperse Blue succinic acid derivative
3. Amide coupling of
4 with the Disperse Blue derivative
3 produced compound
5, and derivatisation of the 3′-hydroxyl group with succinic anhydride gave
6 in good yield. Coupling of compound
6 to amino-functionalised LCAA-CPG gave Disperse Blue CPG
7 suitable for use in automated DNA synthesis (resin loading 27 µmol g
−1). Optimum conditions for deprotection of oligonucleotides containing the Disperse Blue quencher have been determined.
10 |
| | Scheme 2 Synthesis of Disperse Blue quencher CPG.
Reagents and conditions: i, DEAD (1.1 eq.), PPh
3 (1.1 eq.), phthalimide (1.1 eq.), THF, rt, 1.5 h, then hydrazine monohydrate (5 eq.), CH
2Cl
2–MeOH (1 : 1), rt, 15 h, 90% over two steps; ii, succinic anhydride (1.1 eq.), DMAP (0.1 eq.), pyridine, rt, 1.5 h, 90%; iii, DIPEA (6 eq.), HOBT (1.1 eq.), EDC (1.1 eq.),
3 (1.1 eq), DMF, rt, 15 h, 60%; iv, succinic anhydride (1.2 eq.), DMAP (0.1 eq.), pyridine, rt, 18 h then 60 °C, 5 h, 94%; v, LCAA-CPG, EDC (5 eq.), DIPEA (5 eq.), 1% DIPEA in CH
2Cl
2, rt, 4 h, loading = 27 µmol g
−1.
| |
Synthesis and evaluation of Disperse Blue TaqMan® probes
In the Taqman
® method, fluorescence quenching occurs by FRET
13 within the intact probe. During the extension phase of PCR the probe binds to its specific target and is degraded by the 5′→3′ exonuclease activity of
Taq polymerase. This releases the quencher and fluorophore into solution, where they are spatially separated, resulting in an increase in fluorescent signal. TaqMan
® probes targeting a 27-mer sequence in the human β-actin gene (ODN 08 to ODN 15) were synthesised using the 3′-Disperse Blue quencher in combination with a number of different 5′-fluorophores. The Disperse Blue TaqMan
® probes performed significantly better than the methyl red counterparts as quenchers of FAM, JOE, and TAMRA (
Fig. 5). This improvement in performance is manifested as an increase in fluorescent signal, and in the case of TAMRA a decrease in
Ct (cycle number at which the fluorescent signal emerges from the background). TaqMan
® probes containing ROX fluorophore were also studied, but it was not possible to use a passive reference and consequently fluctuations in the PCR curves were observed, as the software was unable to create a flat baseline (data not shown).
 |
| | Fig. 5 Comparison of Disperse Blue and methyl red in a TaqMan
® PCR assay using the fluorophores FAM, JOE and TAMRA. The relative fluorescence
vs. the number of PCR cycles is shown.
| |
Synthesis of Disperse Blue dU phosphoramidite (
10)
The synthesis of fluorogenic oligonucleotide probes sometimes requires the quencher to be incorporated within the oligonucleotide sequence (
e.g. Scorpion primers, certain TaqMan
® probes). In order to fulfil this requirement Disperse Blue must be attached to a nucleoside that is suitable for Watson–Crick base pairing and be converted to a phosphoramidite for incorporation during automated oligonucleotide synthesis. The modified 2′-deoxyuridine nucleoside
814 was chosen and coupled to Disperse Blue succinic acid derivative
3 to give the Disperse Blue 2′-deoxyuridine nucleoside
9. This was then converted to the Disperse Blue phosphoramidite
10 for internal-labelling of oligonucleotides at thymidine sites (
Scheme 3).
 |
| | Scheme 3 Synthesis of Disperse Blue 2′-deoxyuridine phosphoramidite.
Reagents and conditions: i, EDC (1.4 eq.), HOBt (1.4 eq.), DIPEA (1.4 eq.),
3 (0.9 eq.), DMF, rt, 18 h, 45%; ii, 2-cyanoethyl-
N,
N-diisopropyl chlorophosphine (1.5 eq.), DIPEA (4.0 eq.), THF, rt, 2.5 h, 93%.
| |
UV Thermal melting
A series of oligonucleotides (ODN 16-ODN 22 in
Table 1) was synthesised to investigate the effect of the Disperse Blue dU quencher on duplex stability by UV thermal melting. The oligonucleotides (ODN 16–18) contain the Disperse Blue 2′-deoxyuridine nucleoside at three different positions, and ODN 19 is an unmodified control oligonucleotide of the same sequence. Three complementary strands were prepared, all containing the core sequence of ODN 21 (blunt-end complement). ODN 20 differs by the addition of 5′-FAM and ODN 22 has two additional bases (AT) at the 5′-end. In all cases incorporation of the Disperse Blue quencher had a stabilising effect on the duplex, causing an increase in
Tm by up to 6.4 °C (
Table 2).
Table 2 UV-Melting data (all temperatures in °C);
DU = Disperse Blue quencher 2′-deoxyuridine
| Sequence | Oligonucleotide | TmODN20 (5′ FAM complement)
| Δ
Tma | TmODN21 (blunt complement)
| Δ
Tma | TmODN22 (long complement)
| Δ
Tma |
|---|
| Δ
Tm =
Tm of duplex containing quencher −
Tm of unmodified duplex.
|
|---|
| TCCAAGTTTTT(
DU)CTA
| ODN 16 | 56.7 | 5.6 | 55.1 | 3.4 | 56.3 | 4.1 |
| TCCAAG(
DU)TTTTTCTA
| ODN 17 | 54.2 | 3.1 | 55.0 | 3.3 | 55.4 | 3.2 |
| TCCAAGTTTTTTC(
DU)A
| ODN 18 | 57.5 | 6.4 | 55.8 | 4.1 | 57.2 | 5.0 |
| |
| TCCAAGTTTTTTCTA (unmodified) | ODN 19 | 51.1 | — | 51.7 | — | 52.2 | — |
The increase in
Tm was greatest when the quencher was placed close to the end of the duplex (ODN 18) and least when the quencher was in the middle (ODN 17). In the latter case changing the nature of the complementary oligonucleotide had very little effect on
Tm. However, if the complementary sequence was labelled at the 5′-end with FAM there was significant stabilisation when the quencher and the fluorophore were in close proximity. The Disperse Blue moiety protrudes into the major groove from the 5-position of the uracil base, and with Disperse Blue dU:dA as the penultimate base pair in the duplex, the anthraquinone moiety is able to come into close contact with the 5′-FAM. There are several factors that could contribute to the general duplex-stabilising effect of the Disperse Blue dU monomer; the presence of the propyne moiety at the 5-position of the heterocyclic base (which is known to stabilise DNA duplexes), stabilisation from the anthraquinone moiety by intercalation,
15 stacking of the anthraquinone at the 5′- or 3′-end of the duplex to reduce fraying,
16 and/or binding in the minor groove.
17 Factors that stabilise probe:target duplexes can be utilised in genetic analysis to facilitate the use of shorter probes and thereby make it possible to achieve greater discrimination between the wild-type and single point mutations.
Scorpion primer analysis
Stem-loop Scorpion primers consist of a probe sequence held in a hairpin loop conformation by a short complementary stem duplex at the 5′- and 3′-termini of the probe.
3 A fluorophore is attached to the 5′-end of one arm of the stem and a quencher to the 3′-end. The quencher is attached to the 5′-end of a PCR primer
via a PCR stopper. Scorpion primers anneal to the target DNA in the cooling phase of the PCR cycle post-denaturation, and the primer element is extended during the amplification step. The resultant amplicon contains a sequence that is complementary to the probe, which is rendered single-stranded during the denaturation stage of each PCR cycle. On cooling, the probe is free to bind to this complementary sequence, producing an increase in fluorescence, as the quencher is no longer in the vicinity of the fluorophore.
We utilised the W1282X locus
18 of the human ABCC7 gene
19,20 as the Scorpion test system. This gene resides on chromosome 7 and produces the cystic fibrosis transmembrane conductance regulator protein. Specific mutations cause cystic fibrosis, an autosomal recessive disease that is estimated to occur in 1/2000 live births and is carried by 5 percent of the population. These mutations affect the transport of chloride ions across the apical membrane of epithelial cells, producing a number of clinical symptoms.
Two FAM Scorpion primers with either methyl red (ODN 23) or Disperse Blue (ODN 24) quenchers were synthesised targeting a 17-mer wild-type sequence in the W1282X locus. For the Disperse Blue Scorpion the derivatised 2′-deoxyuridine phosphoramidite
10 was used. W1282X is a single point mutation (G to A transition), so the wild-type Scorpions form a duplex with the mutant target that contains a CA mismatch. The Disperse Blue Scorpion gave comparable results to the methyl red Scorpion in terms of
Ct value and fluorescent signal (
Fig. 6), and both Scorpions discriminated clearly between the wild-type (homozygote) and heterozygote.
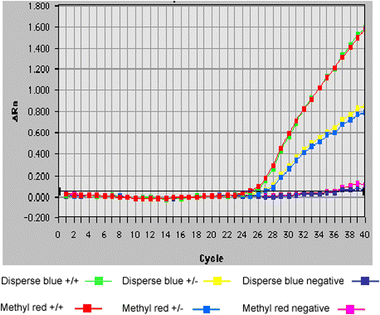 |
| | Fig. 6 Comparison of Disperse Blue and methyl red as quenchers of FAM in a stem-loop Scorpion primer assay. The relative fluorescence
vs. number of PCR cycles is shown.
| |
The alternative “duplex” Scorpion primer format was also evaluated.
4 In this format there is no stem-loop and instead the probe element has a fluorophore attached at its 5′-end, which is annealed to a separate complementary oligonucleotide bearing a quencher at its 3′-end. The mechanism of action is essentially the same as in the stem-loop format, but the quenching mechanism is inter-molecular as opposed to intra-molecular. Two FAM duplex Scorpion primers with methyl red (ODN 25) or Disperse Blue (ODN 26) as quencher were synthesised and compared in real-time PCR on the ABI Prism™ 7700 Genetic analyser (
Fig. 7). The Disperse Blue duplex Scorpion performed better than the methyl red version, both in terms of
Ct and fluorescent signal. Each Scorpion discriminated between homozygote and heterozygote in a cystic fibrosis mutation assay, but the fluorescent signal-to-noise ratio was significantly enhanced with the Disperse Blue quencher.
 |
| | Fig. 7 Comparison of Disperse Blue and methyl red as quenchers of FAM in a duplex Scorpion primer assay. The relative fluorescence
vs. number of PCR cycles is shown.
| |
Studies were also made on the Rotor-Gene genetic analyser (
Fig. 8). This machine allows the longer wavelength Cyanine dyes to be used and so comparison was made between methyl red (ODN 28) and Disperse Blue (ODN 29), with the fluorescent dyes FAM (ODN 30), Cy3 (ODN 31) and Cy5 (ODN 32) in the duplex Scorpion format. Good amplification was seen for both quenchers with the FAM containing complement, although
Ct and fluorescent signal were better for Disperse Blue. The cyanine dye Scorpions only showed good amplification when paired with Disperse Blue. The relatively poor amplification observed with the methyl red/cyanine dye combination is not just a consequence of the improved signal-to-noise ratio produced by Disperse Blue, and further investigations are needed to ascertain the exact reason for this observation. Most importantly Disperse Blue and cyanine dye combinations gave excellent signals relative to background with the greatest fluorescence output seen for Cy5.
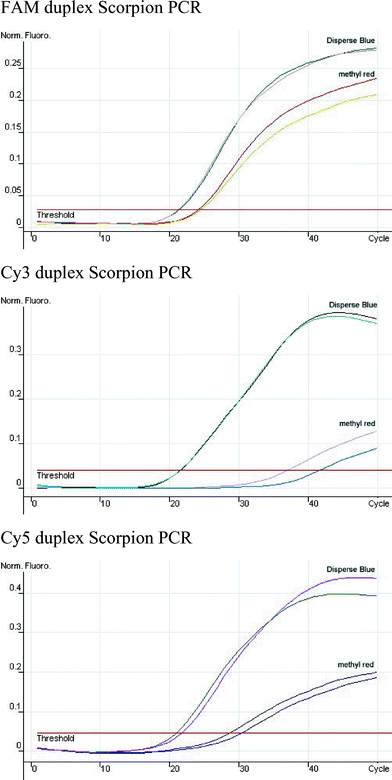 |
| | Fig. 8 Comparison of Disperse Blue and methyl red as quenchers of FAM, Cy3 and Cy5 in a duplex Scorpion primer assay. The normalised fluorescence
vs number of PCR cycles is shown. Each PCR run was carried out in duplicate.
| |
Conclusion
A non-fluorescent quencher based on Disperse Blue has been incorporated into synthetic oligonucleotides at the 5′- and 3′-ends and internally within the DNA sequence. The quencher absorbs over a broad region of the visible spectrum and has been used to quench fluorophores that emit in the 514–704 nm range. The utility of the quencher has been demonstrated in real-time PCR using simple hybridisation probes, TaqMan
® probes and two types of Scorpion primer. In most cases much improved signal-to-noise ratios were observed, particularly with the longer wavelength dyes. The Disperse Blue quencher significantly stabilises DNA duplexes, and this may facilitate the design of shorter, more specific probes.
Experimental
Preparation of compounds
Reagents were purchased from Aldrich, Avocado, Cruachem, Fluka, Lancaster or Link Technologies and used without purification, with the exception of the following solvents that were purified by distillation: methanol (over iodine and magnesium), tetrahydrofuran (over sodium wire and benzophenone), dichloromethane, diisopropylethylamine, pyridine and triethylamine (over calcium hydride). All chemical reactions were carried out under argon using oven dried glassware. Column chromatography was performed under pressure using Fisher Scientific DAVISIL 60A (35–70 micron) silica. Compounds were visualised by irradiation at 254 nm or by staining with anisaldehyde. Thin layer chromatography was performed using Merck Kieselgel 60 F24 (0.22 mm thickness, aluminium backed).
1H NMR spectra were measured at 300 MHz on a Bruker AC300 spectrometer or 400 MHz on a Bruker DPX400 spectrometer.
13C NMR and
31P NMR spectra were measured at 75 MHz and 90 MHz respectively on a Bruker AC300 spectrometer. Chemical shifts are given in ppm relative to tetramethylsilane and
J values are given in Hz. Low-resolution mass spectra were recorded using the electrospray technique on a Fisons VG platform instrument in acetonitrile or a Waters ZMD quadrupole mass spectrometer in methanol or water. High-resolution mass spectra were recorded using the electrospray technique on a Bruker APEX III FT-ICR mass spectrometer in methanol, acetonitrile or water. Infrared spectra were recorded on a BIORAD FT-IR using a Golden Gate adapter and BIORAD WIN-IR software or a Satellite FT-IR using a Golden Gate adapter and WIN FIRST-lite software. Absorptions are described as strong (s) or medium (m). UV-vis data was recorded on a Perkin–Elmer Lambda 2 spectrometer, using 1 mL cuvettes at 25 °C. All extinction coefficients are quoted in parentheses (cm
2 mol
−1). Melting points were measured on a Gallenkamp electrothermal melting point apparatus and are uncorrected.
1-(((2-Cyanoethyl-
N,
N-diisopropylphosphoramidyl)ethyl) amino)-4-(methylamino)anthraquinone (
1)
. Crude 1-((2-hydroxyethyl)amino)-4-(methylamino)anthraquinone (Disperse Blue 3, Aldrich) was purified by silica gel column chromatography eluting with ethyl acetate–hexane (1 : 1). The major fraction (pure Disperse Blue 3) (0.30 g, 1.00 mmol) was dissolved in dry dichloromethane (20 mL) and diisopropylethylamine (0.65 g, 5.20 mmol) was added, followed by 2-cyanoethoxy-(
N,
N-diisopropylamino)chlorophosphine (0.28 g, 1.20 mmol), under argon. The reaction was stirred for 3 h then diluted with ethyl acetate (50 mL), washed with saturated potassium chloride (50 mL), dried (anhydrous sodium sulfate) and evaporated to dryness
in vacuo. Purification by silica gel column chromatography under argon, eluting with ethyl acetate, gave a solid which was dissolved in anhydrous acetonitrile (5 mL) and filtered. This gave the title compound as a blue/purple solid that was sensitive to air (0.32 g, 64%).
1H NMR (CDCl
3): 10.85–10.78 (m, 1H, CH
2N
H), 10.55–10.48 (m, 1H, CH
3N
H), 8.26–8.22 (m, 2H, C
H Ar
6,7), 7.62–7.57 (m, 2H, C
H Ar
5,8), 7.20 (d,
J = 9.5 Hz, 1H, C
H Ar
3), 7.15 (d,
J = 9.5 Hz, 1H, C
H Ar
2), 3.95–3.72 (m, 4H, C
H, C
H2N), 3.59–3.41 (m, 4H, C
H2O), 3.00 (d,
J = 5.2 Hz, 3H, C
H3N), 2.55 (t,
J = 6.6 Hz, 2H, C
H2CN), 1.15–1.05 (m, 12H,
iPr–C
H3);
31P NMR (CDCl
3): 148.96; ES
+/MS: 497.3 (M + H)
+, 519.3 (M + Na)
+
1-((2-Aminoethyl)amino)-4-(methylamino)anthraquinone (
2)
. 1-((2-Hydroxyethyl)amino)-4-(methylamino)anthraquinone (Disperse Blue 3) (0.10 g, 0.33 mmol), phthalimide (0.06 g, 0.37 mmol) and triphenylphosphine (0.10 g, 0.37 mmol) were dissolved in dry tetrahydrofuran (20 mL). To this was added a solution of diethylazodicarboxylate (0.07 g, 0.37 mmol) in dry tetrahydrofuran (2 mL). The reaction was stirred for 1.5 h, evaporated to dryness
in vacuo and the residue was dissolved in dichloromethane–methanol (1 : 2, 20 mL). Hydrazine monohydrate (0.083 g, 1.65 mmol) was added and the mixture was stirred for 15 h, after which the solvent was removed
in vacuo. The blue solid was dissolved in dichloromethane (50 mL), washed with sodium hydroxide (2 M, 2 × 50 mL) and the product was removed from the organic layer by washing with aqueous citric acid (10% w/v, 100 mL). The blue aqueous layer was neutralised (saturated sodium bicarbonate, 2 × 100 mL) and the product extracted by washing with dichloromethane (150 mL). The organic layer was dried (anhydrous sodium sulfate) and evaporated to dryness
in vacuo. Purification by silica gel column chromatography, eluting with dichloromethane–methanol (9 : 1) gave the title compound as a blue amprphous solid (0.09 g, 90%).
1H NMR (CDCl
3): 10.79 (br, 1H, CH
2N
H), 10.51 (br, 1H, CH
3N
H), 8.30–8.20 (m, 2H, C
H Ar
6,7), 7.65–7.55 (m, 2H, C
H Ar
5,8), 7.12 (d,
J = 9.5 Hz, 1H, C
H Ar
3), 7.08 (d,
J = 9.5 Hz, 1H, C
H Ar
2), 3.39–3.28 (m, 4H, C
H2N), 3.00 (d,
J = 5.0 Hz, 3H, C
H3N), 1.32 (br s, 2H, N
H2);
13C NMR (CDCl
3): 182.89, 182.68 (
CO), 147.31, 146.45 (
CNH), 134.89, 134.85 (
C Ar), 132.39, 132.35, 126.43, 126.33, 123.82, 123.33 (
CH Ar), 110.45, 110.31 (
C Ar), 46.31, 37.88 (
CH
2), 29.86 (
CH
3); ES
+/MS: 296.0 (M + H)
+; HRMS (ES
+) for C
17H
17N
3O
2 (M + H)
+: calculated 296.1394, found 296.1388. IR
νmax/cm
−1: 3325 (s), 2916 (m), 2857 (m), 1647 (m), 1561 (s), 1518 (s). UV/vis (CH
2Cl
2): 560 nm (4.3 × 10
3 dm
3 mol
−1 cm
−1), 598 nm (7.9 × 10
3 dm
3 mol
−1 cm
−1), 645 nm (8.5 × 10
3 dm
3 mol
−1 cm
−1).
1-(2-Succinamidoethylamino)-4-(methylamino)anthraquinone (
3)
. Compound
2 (0.09 g, 0.34 mmol), succinic anhydride (0.04 g, 0.38 mmol) and
N-dimethylaminopyridine (0.01 g, 0.09 mmol) were dissolved in dry pyridine (20 mL). The reaction was stirred for 1.5 h, evaporated to dryness
in vacuo, and the product purified by silica gel column chromatography, eluting with dichloromethane–methanol (9 : 1) to give the title compound as a blue solid (0.09 g, 90%).
1H NMR (DMSO-d
6): 10.81 (br, 1H, CH
2N
H), 10.62 (br, 1H, CH
3N
H), 8.36–8.27 (m, 2H, C
H Ar
6,7), 8.30–8.20 (m, 1H, N
HCO), 7.78–7.65 (m, 2H, C
H Ar
5,8), 7.59 (d,
J = 9.4 Hz, 1H, C
H Ar
3), 7.42 (d,
J = 9.4 Hz, 1H, Ar
2), 3.61–3.20 (m, 4H, C
H2N), 3.08 (d,
J = 5.2 Hz, 3H, C
H3N), 2.45 (t,
J = 7.5 Hz, 2H, C
H2CO), 2.34 (t,
J = 7.5 Hz, 2H, C
H2CO);
13C NMR (DMSO-d
6): 180.89, 179.85, 173.07, 170.85 (
CO), 146.07, 145.16 (
CNH), 134.89, 134.85, 131.51, 124.93, 123.77, 123.45 (
CH Ar), 107.85, 107.31 (
C Ar), 54.13, 41.77, 39.28, 29.31 (
CH
2), 28.50 (
CH
3); ES
+/MS: 396.0 (M + H)
+; HRMS (ES
+) for C
21H
21N
3O
5 (M + H)
+: calculated 396.1554, found 396.1545. IR
νmax/cm
−1: 3302 (s), 2921 (m), 1650 (m), 1564 (s). UV/vis (CH
2Cl
2): 561 nm (4.2 × 10
3 dm
3 mol
−1 cm
−1), 595 nm (7.8 × 10
3 dm
3 mol
−1 cm
−1), 642 nm (8.7 × 10
3 dm
3 mol
−1 cm
−1). Mp 179–182 °C.
5′-
O-(4,4′-Dimethoxytrityl)-1′-
O-(6-
N1-(2-((4-(methylamino)anthraquinone)amino)ethyl)succinamidohexyl)-2′-deoxy-
D-ribose (
5)
. Compound
3 (0.10 g, 0.26 mmol), 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (EDC) (0.06 g, 0.29 mmol), 1-hydroxybenzotriazole (HOBt) (0.04 g, 0.29 mmol) were dissolved in
N,
N-dimethylformamide (5 mL) and stirred for 10 min. To this was added 1-(6-aminohexyloxy)-5-(4,4′-dimethoxytrityl)-2-deoxy-α-
D-ribose (compound
4) (0.16 g, 0.29 mmol) in
N,
N-dimethylformamide (5 mL) and diisopropylethylamine (0.5 mL). This solution was then added to the acidic mixture and stirred for 15 h. The solvent was then removed
in vacuo and the solid was redissolved in dichloromethane (100 mL), washed with water (100 mL), saturated sodium bicarbonate (100 mL), dried (anhydrous sodium sulfate) and evaporated to dryness
in vacuo to give the title compound as a blue solid (0.14 g, 60%).
1H NMR (CDCl
3): 10.68–10.62 (m, 1H, N
H), 10.53–10.48 (m, 1H, N
H), 8.23–8.19 (m, 2H, C
H Ar
5,8), 7.62–7.56 (m, 2H, C
H Ar
6,7), 7.39–7.09 (m, 11H, C
H Ar), 6.72 (d,
J = 9.0 Hz, 4H, C
H Ar), 6.69–6.64 (m, 1H, N
H), 6.01–5.96 (m, 1H, N
H), 5.12 (d,
J = 4.5 Hz, 1H,
H1′), 4.16–4.09 (m, 2H,
H3′,
H4′′), 3.70 (s, 6H, OC
H3), 3.69–3.60 (m, 1H, C
HO), 3.49–3.44 (m, 4H, C
H2), 3.33–3.27 (m, 1H, C
HO), 3.12–3.01 (m, 4H, NC
H2, C
H25′), 3.00 (d,
J = 5.0 Hz, 3H, NC
H3), 2.52–2.41 (m, 4H, C
H2CO), 2.15–1.90 (m, 2H,
H2′), 1.50–1.21 (m, 8H, C
H2);
13C NMR (CDCl
3): 182.55, 182.09, 171.93, 171.86 (
CO), 158.31 (
COMe), 146.93, 145.71 (
CNH), 144.66, 135.87, 135.78, 134.36, 134.16 (
C), 132.00, 131.84, 130.06, 129.90, 128.16, 128.00, 127.64 (
CH), 126.60 (
C), 125.89, 123.24, 123.02, 112.95 (
CH), 110.10, 109.68 (
C), 104.17 (
CH
1′), 86.57 (
CH
4′), 85.91 (
C) 73.33 (
CH
3′), 67.20 (
CH
2O), 63.94 (
CH
5′), 55.04 (
CH
3), 45.98, 41.79, 40.79 (
CH
2N), 39.34 (
CH
22′), 31.68, 29.33 (
CH
2), 29.26, 29.27 (
CH
2CO,
CH
3), 26.40, 25.71 (
CH
2); ES
+/MS 913.0 (M + H)
+, 935.0 (M + Na)
+; HRMS (ES
+) for C
53H
60N
4O
10 (M + H)
+: calculated 913.4382, found 913.4364. IR
νmax/cm
−1: 3293 (s), 2927 (m), 2868 (m), 1636 (m), 1572 (s), 1551 (s), 1513 (s). UV/vis (CH
2Cl
2): 562 nm (11.1 × 10
3 dm
3 mol
−1 cm
−1), 595 nm (19.6 × 10
3 dm
3 mol
−1 cm
−1), 642 nm (20.7 × 10
3 dm
3 mol
−1 cm
−1). Mp 150–152 °C.
5′-
O-(4,4′-Dimethoxytrityl)-1′-
O-(6-
N1-(2-((4-(methylamino)anthraquinone)amino)ethyl)succinamidohexyl)-2′-deoxy-3′-
O-oxobutanoic acid-
D-ribose (
6)
. Compound
5 (0.15 g, 0.16 mmol) was dissolved in dry pyridine (5 mL) and succinic anhydride (0.04 g, 0.40 mmol) and 4-(dimethylamino)pyridine (20.00 mg) were added. The mixture was heated at 60 °C for 8 h and the solvent then removed
in vacuo. The residue was dissolved in dichloromethane (150 mL), washed with saturated potassium chloride (150 mL), dried (anhydrous sodium sulfate) and evaporated to dryness. Purification by column chromatography eluting with dichloromethane–methanol–acetic acid (99 : 1 : 0 to 80 : 20 : 0 to 79 : 20 : 1) gave the title product as a blue amorphous solid (0.15 g, 94%).
1H NMR (CDCl
3): 10.50–10.40 (m, 2H, N
H), 8.15–8.08 (m, 2H, C
H Ar
6,7), 7.55–7.49 (m, 2H, C
H Ar
5,8), 7.35–7.06 (m, 11H, C
H Ar), 6.90–6.79 (m, 2H, N
H), 6.71 (d,
J = 8.5 Hz, 4H, C
H Ar), 5.14 (d,
J = 4.5 Hz, 1H,
H1′), 5.07 (d,
J = 6.0 Hz, 1H,
H3′), 4.13–4.09 (m, 1H,
H4′), 3.68 (s, 6H, OC
H3), 3.62–3.33 (m, 4H, C
H2O,
H5′), 3.31–3.11 (m, 4H, NC
H2), 3.12–3.02 (m, 3H, NC
H3), 2.96–2.86 (m, 2H, NC
H2), 2.60–2.42 (m, 8H, C
H2CO), 2.30–1.85 (m, 2H,
H2′), 1.70–1.00 (m, 8H, C
H2);
13C NMR (CDCl
3): 182.55, 182.09, 171.86, 171.93 (
CO), 157.30 (
COCH
3), 145.84, 144.69 (
CNH), 143.69, 134.87, 134.76, 133.23, 132.89 (
C), 130.90, 130.66, 128.91, 127.02, 126.62 (
CH), 125.60 (
C), 124.92, 124.77, 122.25, 121.96, 111.95 (
CH), 110.10, 109.68 (
C), 102.69 (
CH
1′), 84.94 (
CH
4′), 82.09 (
C) 74.42 (
CH
3′), 65.61 (
CH
2O), 62.91 (
CH
5′), 54.04 (
CH
3), 45.98, 41.79, 40.79 (
CH
2N), 39.34 (
CH
22′), 31.68, 29.33 (
CH
2), 29.27, 29.26 (
CH
2CO,
CH
3), 26.40, 25.71 (
CH
2); ES
+/MS: 1034.9 (M + Na)
+, 1051.4 (M + K)
+. IR
νmax/cm
−1: 3322 (s), 2941 (m), 2606 (s), 1655 (s), 1574 (s). UV/vis (CH
2Cl
2): 597 nm (10.6 × 10
3 dm
3 mol
−1 cm
−1), 642 nm (9.7 × 10
3 dm
3 mol
−1 cm
−1).
5′-
O-(4,4′-Dimethoxytrityl)-1′-
O-(6-
N1-(2-((4-(methylamino)anthraquinone)amino)ethyl)succinamidohexyl)-2′-deoxy-3′-
O-oxobutanoic acid-
D-ribose LCAA-CPG (
7)
. All glassware was soaked in trimethylsilyl chloride–dichloromethane (9.5 : 0.5) for 0.5 h and then rinsed with acetone, water, then acetone again, before drying in an oven. The resin (0.50 g) was washed with a solution of dichloromethane–diisopropylethylamine (99 : 1) to liberate the free amines and then dried under argon. Compound
6 (0.05 g, 0.05 mmol) was dissolved in dichloromethane–diisopropylethylamine (99 : 1) (0.5 mL) and 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (47.00 mg, 0.25 mmol) was added. The resin (long-chain alkylamino controlled pore glass, Link Technologies, 0.50 g) was added and the mixture agitated gently for 2 h. The resin was filtered and washed with dichloromethane–diisopropylethylamine solution (99 : 1, 3 × 10 mL), soaked in a mixture of acetic anhydride, 2,6-lutidine and 1-methylimidazole in tetrahydrofuran for 1 h (DNA synthesis capping agents), washed with dichloromethane–diisopropylethylamine (99 : 1, 3 × 10 mL) solution and finally with diethyl ether (3 × 20 mL). A small sample of the resin (2 mg) was dissolved in conc. hydrochloric acid–ethanol (3 : 2 v/v, 25 mL) and shaken for 5 min. The absorbance of the orange solution (1 mL, 1 cm path length) was measured at 495 nm (abs = 0.156) and the loading of the resin was then calculated as 27 µmol g
−1 from the known absorbance of 1 µmol of DMT cation in 25 mL volume (71.7). Columns were prepared for 0.2 µmol scale oligonucleotide synthesis by adding 9 mg of functionalised resin to each column (Glen Research screw-fit columns).
5′-
O-(4,4′-Dimethoxytrityl)-5-(3-(6-
N1-(2-((4-(methylamino)anthraquinone)amino)ethyl)succinamidohexamide)propynyl)-2′-deoxyuridine (
9)
. Compound
3 (0.3 g, 0.78 mmol), 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide hydrochloride (0.23 g, 1.2 mmol) and 1-hydroxybenzotriazole (0.16 g, 1.2 mmol) were dissolved in dry
N,
N-dimethylformamide (20 mL) and stirred for 10 min. To this was added 6-amino-
N-(3-(5′-(4,4′-dimethoxytrityloxy)-2′-deoxyuridin-5-yl)prop-2-ynyl)hexanamide
8 (0.6 g, 0.87 mmol) and diisopropylethylamine (0.16 g, 1.2 mmol) in dry
N,
N-dimethylformamide (10 mL). The reaction mixture was stirred at room temperature for 18 h then the solvent was removed
in vacuo. The residue was dissolved in dichloromethane (150 mL), washed with water (150 mL), saturated sodium bicarbonate (150 mL), dried with sodium sulfate, evaporated to dryness and purified by column chromatography with silica gel (dichloromethane–methanol, 9 : 1) to yield the title compound (0.38 g, 45%) as a blue solid.
1H NMR (CDCl
3): 10.64 (s, 1H, CH
2N
H), 10.50 (br, 1H, CH
3N
H), 8.16–8.07 (m, 2H, C
H Ar
6,7), 7.98–7.93 (m, 1H, N
H), 7.55–7.45 (m, 2H, C
H Ar
5,8), 7.40–7.05 (m, 12H, C
H Ar, N
H), 7.00–6.87 (m, 2H, C
H Ar), 6.72 (d,
J = 8.5 Hz, 4H, C
H Ar), 6.35 (s, 1H, N
HCO), 6.18 (t,
J = 6.5 Hz, 1H,
H1′), 4.49–4.41 (m, 1H,
H3′), 4.09–3.96 (m, 1H,
H4′), 3.90–3.70 (m, 2H, C
H2), 3.65 (s, 6H, C
H3O), 3.42–3.31 (m, 4H, C
H2N), 3.29–3.16 (m, 2H,
H5′), 3.12–2.99 (m, 2H, C
H2), 2.87 (d,
J = 5.0 Hz, 3H, C
H3N), 2.45–2.31 (m, 4H, C
H2CO), 2.25–1.85 (m, 4H, C
H2CO,
H2′), 1.56–1.12 (m, 6H, C
H2);
13C NMR (CDCl
3): 181.73, 181.29, 172.90, 172.34, 172.23, 162.70 (
CO), 158.14 (
C), 149.53, 146.59, 145.50 (
CNH), 144.18 (
C), 142.48 (
CH), 135.18, 133.97, 133.73 (
C), 131.56, 131.45, 129.59, 127.60, 127.53, 126.52, 125.61, 125.50, 122.78, 112.79 (
CH Ar), 109.42, 109.04 (
C Ar), 98.96 (
C), 89.05 (
C), 86.49 (
C), 86.21 (
CH), 85.39 (
CH), 73.92 (
C), 71.49 (
CH), 63.33 (
CH
2), 54.86 (
CH), 52.50 (
CH
2), 41.48, 41.25, 39.06, 38.82, 35.39, 31.35, 29.41 (
CH
2), 28.96 (
CH
3), 28.51, 25.71, 24.51 (
CH
2); ES
+/MS: 1096.5 (M + Na)
+; HRMS (ES
+) for C
60H
63N
7O
12 (M + H)
+: calculated 1074.4636, found 1074.4607. IR
νmax/cm
−1: 3317 (s), 3073 (m), 2941 (m), 1650 (s), 1574 (m). UV/vis (CH
2Cl
2): 560 nm (6.3 × 10
3 dm
3 mol
−1 cm
−1), 596 nm (12.2 × 10
3 dm
3 mol
−1 cm
−1), 644 nm (13.6 × 10
3 dm
3 mol
−1 cm
−1). Mp 138–144 °C.
5′-
O-(4,4′-Dimethoxytrityl)-5-(3-(6-
N1-(2-((4-(methylamino)anthraquinone)amino)ethyl)succinamidohexamide)propynyl)-3′-
O-(2-cyanoethyl-
N,
N-diisopropylphosphoramidyl)-2′-deoxyuridine (
10)
. To compound
9 (0.35 g, 0.33 mmol) in dry dichloromethane (30 mL) was added diisopropylethylamine (0.23 mL, 1.32 mmol) and 2-cyanoethyl-
N,
N-diisopropyl chlorophosphine (0.113 mL, 0.50 mmol). The reaction mixture was stirred under argon for 2.5 h then concentrated
in vacuo. The residue was dissolved in ethyl acetate (degassed with argon), washed with saturated potassium chloride, dried (anhydrous sodium sulfate), reduced to dryness
in vacuo then purified by silica gel column chromatography under argon to yield the title product as an oil (0.36 g, 93%).
1H NMR (CDCl
3): 10.73 (s, 1H, CH
2N
H), 10.61–10.48 (m, 1H, CH
3N
H), 8.31–8.15 (m, 2H, C
H Ar
6,7), 8.10–7.98 (m, 1H, N
H), 7.64–7.50 (m, 2H, C
H Ar
5,8), 7.41–7.01 (m, 14H, C
H Ar, N
H), 6.78 (d,
J = 8.5 Hz, 4H, C
H Ar), 6.61 (s, 1H, N
HCO), 6.26 (t,
J = 6.5 Hz, 1H,
H1′), 4.59–4.48 (m, 1H,
H3′), 4.27–4.16 (m, 1H,
H4′), 4.15–3.90 (m, 2H, C
H2), 3.90–3.21 (m, 16H, C
H3O, C
H2N,
iPr
H, C
H2O), 3.19–3.08 (m, 2H, C
H2N,
H5′), 2.98 (d,
J = 5.0 Hz, 3H, C
H3N), 2.65–2.41 (m, 6H, C
H2), 2.38–1.88 (m, 2H, C
H2O,
H2′), 1.61–0.97 (m, 18H, C
H2,
iPrC
H3); ES
+/MS: 1296.5 (M + Na)
+.
31P NMR (CDCl
3): 149.60, 149.28.
Oligonucleotide synthesis
All oligonucleotides were synthesised on an ABI 394 DNA synthesiser by standard automated solid-phase methods using β-cyanoethyl phosphoramidite chemistry (0.2 µmol scale). All coupling efficiencies (as measured by automated trityl analysis) were >98%. For the synthesis of Cy3, Cy5, Cy5.5 and Disperse Blue oligonucleotides, fast-deprotecting dmf dG and acetyl dC phosphoramidites were used (Link Technologies Ltd.). Disperse Blue-functionalised resin was packed into ABI 394 DNA synthesis columns (Glen Research Inc.). Deprotection of oligonucleotides containing the Disperse Blue quencher was carried out by transferring the resin-bound oligonucleotides from the columns into 3 mL vials and treating with water–methanol–
tert-butylamine (2 : 1 : 1) at 70 °C for 2.5 h. Oligonucleotides containing cyanine dyes were deprotected in conc. aqueous ammonia at 55 °C for 1 h only, and all other oligonucleotides were cleaved and deprotected using concentrated aqueous ammonia at 55 °C for 5 h. Methyl red aminohexyl deoxyribose CPG and FAM aminohexyl deoxyribose CPG were used to add 3′-MeRed and 3′-FAM respectively and were prepared by published methods.
9 All fluorescent dyes except ROX, TAMRA, and JOE were added as phosphoramidites during oligonucleotide synthesis. Oligonucleotides with 3′-cyanine dyes were synthesised on a thymidine column and the first addition during solid-phase synthesis was the cyanine dye. This method was used due to the inavailability of cyanine-dye CPG. ROX, TAMRA and JOE NHS esters (1 mg in DMSO, 25 µL) were added post-synthetically to the 5′-aminohexyl- or 3′-aminohexyldeoxyribose-functionalised oligonucleotides which were made from the corresponding aminolink monomer or CPG.
9 The amino-modified oligonucleotides were labelled by dissolving in 0.5 M sodium bicarbonate/carbonate buffer at pH 9.25 (60 µL) and the resultant solution was set aside overnight at room temperature. Excess dye was removed by Sephadex gel-filtration (Nap 10, Pharmacia), and oligonucleotides were purified by reversed-phase Gilson HPLC on a C8 (octyl) column, eluting with a gradient of 0% to 40% acetonitrile in 0.1 M ammonium acetate buffer.
21TaqMan
® real-time PCR
TaqMan
® PCR reactions were carried out on an ABI Prism™ 7700 Genetic analyser. All reagents were supplied by Eurogentec. Total reaction volume was 50 µL and the reaction mix contained 1 × reaction buffer with ROX passive reference to normalise the data. The ABI Prism™ 7700 uses ROX as a passive reference to normalise the data and account for pipetting inaccuracies between samples. No passive reference was used when testing the ROX labelled probe and consequently baseline fluctuations were observed, as the software was unable to create a flat baseline (data not shown). The reaction mix also included a standard dNTP mix (200 µM, including dUTP), 4 mM MgCl
2, 1 unit of HotGoldStar™
Taq polymerase and sterile water. Primers and probes were used at a concentration of 0.5 µM and were designed to detect human β-actin.
22 Human genomic DNA (30 ng, Roche) was added to each reaction and this was replaced with sterile water for negative controls, run with each set of samples. Standard TaqMan
® PCR cycling conditions were used, namely 95 °C for 10 min (initial denaturation), then 40 cycles of 95 °C for 15 s, 60 °C for 1 min.
Scorpion primer real-time PCR
The Scorpion sequences used to evaluate the fluorescence quenchers (
Table 1) were targeted at a 17- mer region of the wild-type W1282X locus of the human ABCC7 gene.
3,4 The sequence of the reverse primer was 5′-GGCTAAGTCCTTTTGCTCAC-3′. Total reaction volumes were 25 µL, containing 12.5 ng of Human genomic DNA (NA 11472 (wild-type) and NA 1723 (W1282X heterozygote) purchased from Coriell). DNA was replaced with sterile water for the negative controls. Concentrations of reagents were as follows: 1 × PCR buffer containing ROX passive reference (Eurogentec), 200 µM dNTPs (a mix of dCTP, dGTP, dATP, dTTP and dUTP), 0.5 µM Scorpion primer and reverse primer, 2.5 µM quencher oligonucleotide for the duplex Scorpion, 0.5 units of HotGoldStar polymerase (Eurogentec), 4 mM magnesium chloride. All Scorpion PCR reactions were carried out on an ABI PRISM™ 7700 Genetic analyser with the quencher option turned off. Cycling was as follows: 95 °C for 10 min (initial denaturation), then 40 cycles of 95 °C for 30 s, 53 °C for 30 s, 72 °C for 30 s. Fluorescence was monitored during the 53 °C step.
The duplex Scorpions containing the cyanine dyes (
Fig. 8) were studied on a Corbett Research Rotorgene Rg-3000. The PCR curves were quantitised and show the relative fluorescence with respect to the background. These PCRs were carried out with 2 mM MgCl
2, 2U Taq Polymerase, 0.5 µM primers and 2.5 µM quencher. Cycling conditions were: 95 °C for 5 min, then 50 cycles of 95 °C for 30 s, 40 °C for 30 s, 72 °C for 30 s.
Spectrometric measurements
Fluorescence spectra were recorded on a PE LS50B luminescence spectrometer with 5 nm slit widths using quartz glass 3 mL cuvettes. All hybridisation assays were performed in buffer consisting of sodium phosphate (100 mM), EDTA (1 mM) and sodium chloride (100 mM), pH 7.0. Fluorescent dyes were excited at their absorption maxima: FAM (495 nm), Cy3 (550 nm), TAMRA (555 nm), Cy5 (650 nm), Cy5.5 (690 nm) and emission intensity measured at the experimentally determined maxima: FAM (514 nm), Cy3 (564 nm), TAMRA (584 nm), Cy5 (666 nm), Cy5.5 (704 nm). UV/vis data were recorded on a Perkin–Elmer Lambda 2 spectrometer, using 1 mL cuvettes at 25 °C. UV thermal melting was performed on a Perkin–Elmer Lambda 2 UV/vis spectrometer connected to a PTP-1 temperature programmer, and data acquisition was recorded using the PECSS software. All UV melts were performed in triplicate.Acknowledgements
Jonathan May was funded by an EPSRC CASE studentship with Oswel Research Products Ltd., and Ian van Delft was the recipient of an EPSRC Analytical Chemistry PhD studentship.References
- D. Whitcombe, S. Kelly, J. Mann, J. Theaker, C. Jones and S. Little, Am. J. Human Genet., 1999, 65, 2333 Search PubMed.
- D. Whitcombe, J. Theaker, S. P. Guy, T. Brown and S. Little, Nat. Biotechnol., 1999, 17, 804–807 CrossRef CAS.
- N. Thelwell, S. Millington, A. Solinas, J. A. Booth and T. Brown, Nucleic Acids Res., 2000, 28, 3752–3761 CrossRef CAS.
- A. Solinas, L. J. Brown, C. McKeen, J. M. Mellor, J. Nicol, N. Thelwell and T. Brown, Nucleic Acids Res., 2001, 29, e96 CrossRef CAS.
- S. Tyagi and F. R. Kramer, Nat. Biotechnol., 1996, 14, 303–308 CrossRef CAS.
- S. Tyagi, D. P. Bratu and F. R. Kramer, Nat. Biotechnol., 1998, 16, 49–53 CrossRef CAS.
- K. J. Livak, S. A. J. Flood, J. Marmaro, W. Giusti and K. Deetz, PCR Methods Appl., 1995, 357 Search PubMed.
- P. M. Holland, R. D. Abramson, R. Watson and D. H. Gelfand, Proc. Natl. Acad. Sci. U. S. A., 1991, 88, 7276–7280 CAS.
- C. M. McKeen, L. J. Brown, J. T. G. Nicol, J. M. Mellor and T. Brown, Org. Biomol. Chem., 2003, 1, 2267–2275 RSC.
- J. P. May, L. J. Brown, I. Rudloff and T. Brown, Chem. Commun., 2003, 970–971 RSC.
- L. J. McBride and M. H. Caruthers, Tetrahedron Lett., 1983, 24, 245–248 CrossRef CAS.
- S. L. Beaucage and M. H. Caruthers, Tetrahedron Lett., 1981, 22, 1859–1862 CrossRef CAS.
- P. Wu and L. Brand, Anal. Biochem., 1994, 218, 1–13 CrossRef CAS.
- L. J. Brown, J. P. May and T. Brown, Tetrahedron Lett., 2001, 42, 2587–2591 CrossRef CAS.
- B. Armitage, T. Koch, H. Frydenlund and H. Orum, Nucleic Acids Res., 1998, 26, 715–720 CrossRef CAS.
- D. G. Norman, R. J. Grainger, D. Uhrin and D. M. J. Lilley, Biochemistry, 2000, 39, 6317–6324 CrossRef CAS.
- P. B. Dervan, Science, 1986, 232, 464–471 CrossRef CAS.
- B.-S. Kerem, J. Zielenski, D. Markiewicz, D. Bozon, E. Gazit, J. Yahav, D. Kennedy, J. R. Riordan, F. S. Collins, J. M. Rommens and L.-C. Tsui, Proc. Natl. Acad. Sci. U. S. A., 1990, 87, 8447–8451 CAS.
- J. M. Rommens, M. C. Iannuzzi, B.-S. Kerem, M. L. Drumm, G. Melmer, M. Dean, R. Rozmahel, J. L. Cole, D. Kennedy, N. Hidaka, M. Zsiga, M. Buchwald, J. R. Riordan, L.-C. Tsui and F. S. Collins, Science, 1989, 245, 1059–1065 CrossRef.
- J. Zielenski, R. Rozmahel, D. Bozon, B. Kerem, Z. Grzelczak, J. R. Riordan, J. Rommens and L.-C. Tsui, Genomics, 1991, 10, 214–228 CrossRef CAS.
- T. Brown and D. J. S. Brown, Methods Enzymol., 1992, 211, 20–35 CAS.
- S. A. Bustin, J. Mol. Endocrinol., 2000, 25, 169–193 Search PubMed.
|
| This journal is © The Royal Society of Chemistry 2005 |
Click here to see how this site uses Cookies. View our privacy policy here. 










