Hot off the Press
In the Hot off the Press section of Molecular BioSystems members of the Editorial Board and their research groups highlight recent literature for the benefit of the community. This month the highlighted topics include the intracellular localization of antisense nucleotides, an aptamer-based biosensor, functional proteomics of glycosidases, and a new technique for finding transcription factor binding sites.
Functional proteomics of glycosidases
Recent advances in chemical proteomics have significantly increased the repertoire of tools to assign function to eukaryotic and prokaryotic gene products. Nevertheless, the size and complexity of the proteome continuously challenges postgenomic researchers to develop methodologies to characterize the biology in unique subsets of the proteome. Toward this end, Stephen Withers and his colleagues at the University of British Columbia outline an experimental platform for the identification of active glycosidases in complex proteomes.The challenge of specifically targeting members of this enzyme class is addressed by coupling aspects of activity-based protein profiling and mass spectrometry (MS) technologies. Initially, a mechanism-based inhibitor, 2-deoxy-2-fluoro-xylobioside, was adapted with an electrophile and shown to irreversibly inactivate two purified β-1,4-glycanases via a conserved, catalytic glutamate nucleophile. A pendant biotin moiety was then shown to enrich the covalently-modified active site peptides from an artificial background proteome. Subsequent reduction of a disulfide linker and MS analysis of the probe–peptide conjugate demonstrates that the sequence differences in the residues flanking the conserved glutamate are sufficient “fingerprints” to assign the identity of each β-1,4-glycanase.
A significant advance in the study is the use of the methodology to discover a novel β-1,4-glycanase from the secreted proteome of the soil bacterium Cellulomonas fimi. Notably, although cloning the novel glycanase gene revealed 70% active site domain sequence homology to known xylanases from Streptomyces sp., the enzyme was nevertheless distinguishable by MS analysis of the enriched six-mer active site peptide sequence.
Consequently, a resonant message from this contribution is that high-content functional profiling of complex proteomes can proceed with methodologies designed to globally characterize specific protein classes, thereby rendering tractable the daunting task of assigning function to all gene products.
Omid Hekmat, Young-Wan Kim, Spencer J. Williams, Shouming He, Stephen G. Withers, J. Biol. Chem., 2005, DOI: 10.1074/jbc.M508434200
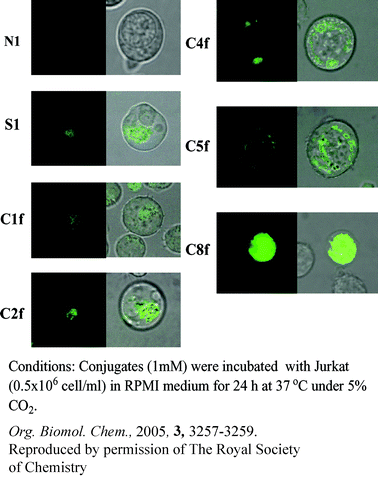
Reviewed by: Michael J. Evans, The Scripps Research Institute, California, USAIntracellular localization of antisense oligonucleotides
When research on antisense oligonucleotides first appeared, a novel type of oligonucleotide therapy was hoped to cause the next big revolution in the pharmaceutical industry. However, the problem of cellular uptake and localization of these oligos remained the main obstacle in making use of their high inhibitory effects.Until now, various gene delivery agents have been used for transport, but often caused cell death and suffered from a lack of precise localization control. Masayuki Fujii and coworkers recently reported oligonucleotide–peptide conjugates which can be used to introduce antisense oliogonucleotides to human leukemia cells.
Several peptide–oligonucleotide conjugates, each containing different signal peptides, were synthesized by solid phase fragment condensation. The oligonucleotides were complementary to the RNA template of human telomerase and signal peptides derived from nuclear export signal (NES) sequence of HIV-1 rev protein and nuclear localization signals (NLS) of SV40 T antigen, influenza virus and HIV-1 tat protein. Additionally, a fluorescent marker was added to each of the conjugates to follow the cellular uptake and localization by confocal laser fluorescence microscopy. As expected, all of the conjugates containing NLS were localized into the nucleus, whereas the ones containing NES remained in the cytoplasm. The control, having no peptide at all, did not show any cellular uptake.
Following the localization, the inhibition of human telomerase elongation was evaluated by telomerase repeat amplification protocol (TRAP). The results showed that inhibition was dramatically affected by localization. One of the oliognucleotide–NLS conjugates completely suppressed telomerase activity (99.6% in 24 hours) indicating that cleverly designed antisense oligonucleotide–peptide conjugates could indeed be used as genetic drugs. It remains only to be seen if and how the cells are affected by introduction of such large unnatural molecules.
T. Kubo, Z. Zhelev, B. Rumiana, H. Ohba, K. Doi, M. Fujii, Org. Biomol. Chem., 2005, 3, 3257–3259
Reviewed by: Ljiljana Fruk, Universität Dortmund, GermanyStreamlining the identification of targets of E3 ligases
Ubiquitin E3 ligases are the “matchmakers” that mediate the coupling of activated ubiquitin derivatives to specific protein substrates. A rate-limiting step in better understanding ubiquitin-affected pathways has been the difficulty of matching particular E3 ligases (of which there are hundreds) to their substrates. However, Daniela Rotin and colleagues recently developed an in vitro luminescence-based assay, which they call AlphaScreen™ technology.This system is based on anti-GST acceptor beads recognizing purified GST-tagged proteins and streptavidin donor beads distinguishing biotinylated ubiquitin (b-Ub). These reagents are incorporated into an in vitro ubiquitylation assay that utilizes purified enzymes, ATP, b-Ub and GST-tagged protein. A correct protein substrate will become ubiquitylated and facilitate a platform for both beads to bind, resulting in the beads being close enough together to generate a luminescent signal when irradiated at 680 nm.
This technology was validated using the already-characterized E3 ligase Rsp5 and 188 purified GST fusion proteins. The data generated from the screen not only identified several known Rsp5 substrates correctly, but also discovered seven novel Rsp5 target proteins. Furthermore, these experiments reveal that this method is greater than 200 times more sensitive than traditional ubiquitylation assays using Western blotting. While this method cannot distinguish between mono- and poly-ubiquitylation, it is a significant advance over current technology.
Bart Kus, Aaron Gajadhar, Karen Stanger, Rob Cho, Warren Sun, Nathalie Rouleau, Tammy Lee, Donovan Chan, Cheryl Wolting, Aled Edwards, Roger Bosse, and Daniela Rotin, J. Biol. Chem., 2005, 280(33), 29470–29478
Reviewed by: Melissa O'Neal, UT-Southwestern Medical Center, USADesigning a biologically active mimic of a peptide that stimulates cell migration
Wounds that heal slowly, or not at all, afflict many people and pose a significant medical challenge. Wound healing is a complex process coordinating a variety of cellular activities such as migration to the wound site, proliferation, remodelling of the extracellular matrix, and vascularization. A collection of polypeptide factors orchestrates the normal process, directing these cellular behaviors at the right time and place. Attempts to accelerate this process by applying relevant polypeptides to the wound site, while showing promise in model studies, have had little clinical success. A possible explanation for failure of this approach is instability of polypeptides in the wound environment. Thus, biologically stable peptidomimetic compounds that stimulate the cellular behaviors involved in wound healing might be valuable therapeutic agents. This possibility motivated efforts by Rodolfo Marquez and co-workers to design and synthesize a peptidomimetic of the tripeptide IGD (isoleucine, glycine, aspartate).This tripeptide motif is a highly conserved element of migration stimulating factor (MSF), which is expressed during wound healing and stimulates several cellular responses that are operative in the wound healing process. These include cell migration, angiogenesis, and production of an extracellular matrix material, hyaluronan. Importantly, the synthetic tripeptide by itself displays biological activity similar to MSF.
The starting point for design of the peptidomimetic was an analysis of the peptide backbone dihedral angles in two NMR-derived solution structures of different domains of MSF. From this analysis, the authors concluded that a benzodiazepine ring system provides a structural framework that looks like the backbone of the tripeptide. Further modelling suggested substituents on the ring system to emulate the isoleucine and aspartate side chains. The authors synthesized the compound that came out of this design process and tested its ability to stimulate fibroblast migration in collagen gel in vitro. The compound had chemokinetic activity in this assay with a dose–response similar to the tetrapeptide IGDS, whereas a control peptide with the reverse sequence, SDGI, showed no activity.
An assessment of the efficacy of the peptidomimetic in the full range of biological activities displayed by IGD and MSF will be an important next step in the development of this compound. In this regard, it is noteworthy that the dose–response curve of the peptidomimetic is similar to that of the tetrapeptide, but its profile does differ significantly, with activity extending to concentrations several-fold higher. This difference suggests some distinction in the signalling mechanisms of the two agents. In any case, this compound is an interesting biologically active lead. The authors indicate that they are developing syntheses of non-racemic versions of the compound (the reported synthesis produces a racemic mixture) and other derivatives, and these second generation compounds may lead to new or improved biological activities. Ultimately, it will be exciting to test these compounds for their effects in vivo, especially their effects on wound healing.
Natalia Shpiro, Ian R. Ellis, Trevor J. Dines, Ana M. Schor, Seth L. Schor, David G. Norman and Rodolfo Marquez, Mol. BioSyst., 2005, DOI: 10.1039/b509023g
Reviewed by: Kevin J. Luebke, UT-Southwestern Medical Center, USACofactor precursors thought to exist in interstellar ice clouds
With the successful Mars Rovers and Cassini missions, the interest in space research is increasing again. Recent work by a group of French, German and Dutch scientists brings us a little closer to understanding what is really going in interstellar space.The team simulated the physical conditions at low pressure, low temperature and UV radiation that may exist in interstellar clouds to see whether it is possible to find any enzyme cofactor precursors in the irradiated samples after warming up to room temperature. The interstellar clouds contain dust particles with an ice mantle containing mostly H2O and then CH3OH, CO, CO2 and NH3. The ice layer of the same composition was prepared in the laboratory and irradiated with UV light. Previously it was shown that building blocks of proteins and nucleic acids were generated under prebiotic conditions but the data on cofactors is rare.
Their results show that hexahydro-1, 3, 5-triazine and its derivatives together with some monopyrrolic molecules are formed in ice clouds under low pressure, low temperature and UV radiation. All of them are precursors of uroporphyrinogen which can be transformed into biological porphyrinoid cofactors.
To exclude the possibility of sample contamination, experiments using 13C-labelled gases were also performed and showed similar composition. These results support the heterotrophy origin theory of the biological cofactors, which assumes that the structural prototypes of cofactors were synthesised under prebiotic conditions and further perfected by organisms. The other, autotrophy origin theory, suggests that organisms are capable of synthesising all carbon constituents directly from CO2.
The researchers are now waiting for the results of the Stardust and Rosseta missions which are going to analyze cometary materials to compare them with their own findings. Stardust should bring the samples of the dust surrounding comet 81p/Wild2 to the Earth, while Rosetta is going to perform in situ GC-MS measurements of comet nucleus. A comparison could shed more light on still unanswered question about the emergence of the first living organisms on Earth.
U. J. Meierhenrich, G. M. Munoz Caro, W. A. Schutte, W. H.-P. Thiemann, B. Barbier, A. Brock, Chem. Eur. J., 2005, 11, 4895–4900
Reviewed by: Ljiljana Fruk, Universität Dortmund, GermanyOligonucleotide fishing: exploring for transcription factor binding sites
High-throughput mapping of DNA–protein contacts is fundamental to understanding coordinated gene expression in eukaryotic cells. Some high-throughput, genome-wide methods, such as ChIP on Chip, DIP-Chip and protein binding microarrays have been applied to this problem. However these microarray-based experiments are expensive, technically demanding, and rely on the availability of high quality antibodies or purified proteins. In order to circumvent some of these limitations, Scot Wolfe and his colleagues describe a modified version of the yeast one-hybrid technique in which a protein–DNA interaction is coupled to the expression of a growth-essential reporter gene.A library of DNA sequences was cloned upstream of reporter genes HIS3 and URA3. They then fused the DNA-binding domain of the transcription factor of interest with the alpha subunit of RNA polymerase. These two constructs were then transformed into a his3 strain of bacteria. Interaction between the transcription factor DNA-binding domain-RNA polymerase fusion and the DNA-reporter gene fusion would activate transcription from the reporter genes, HIS3 and URA3. The inclusion of the URA3 marker, the expression of which can be selected against in a HIS3 strain by including 5-fluoroorotic acid in the media, allowed for negative selection of DNA baits that reported high background arising from what is referred to as self-activation. Colonies positive for HIS3 but negative for URA3 were then selected and sequenced. The conserved motifs are then discovered using the MEME motif finding algorithm.
To validate their system, they used it to probe the binding preferences of Zif268, PLAG1, ZNFp53, Dorsal, LAG-1, Paired, Bgb and Runt, transcription factors representing diverse structural families. Running the sequences of the positive clones through the motif finder algorithm successfully identified the known recognition sites of these proteins.
Having proven that their system works, the team then employed the system, combined with a bioinformatics analysis of the results, to discover binding sites of the odd-skipped protein in Drosophila. These were validated using gel shift and competitive gel shift assays as well as ectopic expression assays in flies. Despite several potentially significant limitations, such as the possible lack of post-translational modifications in bacteria and the likely failure of some factors to express in E. coli, the affordable and straightforward nature of this technology make it useful as a primary tool for hunting for binding sites of transcription factors.
X. Meng, M. H. Brodsky and S. A. Wolfe, Nat. Biotechnol., 2005, 23, 988–94
Reviewed by: Devanjan Sikder, UT-Southwestern Medical Center, USAAptamer based biosensor
Carbon nanotubes and semiconducting nanowires are currently the most widely used one dimensional nanostructures for the development of biosensors. Commonly they are employed as active transducers when the protein recognition element, usually an enzyme or antibody, is used in detection of different targets such as small molecules, proteins or viruses. However, proteins, although reliable recognition probes, are often not robust and synthetically feasible. This leads to high cost and low shelf life of such devices. Now scientists from Korea might bring us a step closer to the commercialization of carbon nanotube biosensors. Jeong-O Lee and colleagues based their biosensor design on the aptamer-modified nanotube. Aptamers are artificial oligonucleotides (RNA or DNA) which can specifically bind a variety of species, often with affinity higher than those of antibodies.15-mer DNA aptamer that binds thrombin was chosen as a model system to investigate the possibility of using aptamers as molecular recognition elements. Using carbodiaimidazole activated Tween-20 as a linker, DNA was immobilized onto vapor deposited carbon nanotubes. Then the thrombin was added to the chip and the conductance measured after the addition showed an abrupt decrease. However, when the controls were used (one without thrombin and one with the protein elastase) no conductance changes were observed. Down to 300 nM of thrombin could be detected, although the team believe that the lowest detection limit of the sensor can be additionally improved.
It is also important to note that all the experiments were performed on the same sensor chip because the removal of protein is easily achieved by washing with guanidine hydrochloride solution. Consequently, in addition to the low cost, fast response, high sensitivity and simplicity of production, the aptamer-based carbon nanotube chips are also recyclable. The researchers mention that they are currently working on the improvement of the sensitivity by using a high performance nanotube file effect transistor and future developments can definitely be expected with great interest.
H.-M. So, K. Won, Y. H. Kim, B.-K. Kim, B. H. Ryu, P. S. Na, H. Kim, J.-O Lee, J. Am. Chem. Soc., 2005, 127, 11906–11907
Reviewed by: Ljiljana Fruk, Universität Dortmund, Germany| This journal is © The Royal Society of Chemistry 2005 |