Immobilization of enzymes on a microchannel surface through cross-linking polymerization†
Takeshi
Honda
a,
Masaya
Miyazaki
*ab,
Hiroyuki
Nakamura
a and
Hideaki
Maeda
*ab
aNanotechnology Research Institute, National Institute of Advanced Industrial Science and Technology (AIST), 807-1 Shuku, Tosu, Saga 841-0052, Japan. E-mail: m.miyazaki@aist.go.jp; maeda-h@aist.go.jp; Fax: +81(942)81-3657; Tel: +81(942)81-3676
bDepartment of Molecular and Material Science, Interdisciplinary Graduate School of Engineering Sciences, Kyushu University, 6-1 Kasuga Koen, Kasuga, Fukuoka 816-8580, Japan
First published on 19th September 2005
Abstract
A novel and facile method for the preparation of an enzyme-immobilized microreactor has been developed in which enzymes are immobilized as an enzyme-polymer membrane formed on the inner wall of the microchannel by a cross-linking polymerization method; the resulting microreactor shows excellent reaction performance and stability against denaturating agents.
Microreactors are potentially powerful tools in the fields of chemistry and biotechnology.1 The excellent performance of microreaction systems is achieved by rapid heat and mass transfers, and larger surface/interface area. These attractive features are favorable to catalytic reactions through catalysts immobilized on the reactor.2 Enzyme catalysts are a catalyst type useful in substance production and with high potential for analytical applications. There is demand for enzymatic microreaction devices in several fields, especially for bioanalyses.3 Several enzyme-immobilized microreactors have been developed. We have developed modified sol-gel techniques for the surface modification of microchannels and their application for the preparation of a microreactor.4 Although it is necessary to reduce cost, effort, and time for production of microreactors, the developed methods of microreactor preparations require high-level techniques and/or multi-step procedures which could lead to low cost performance. Also, disposability of the reactor is considered essential in bioanalyses. The present study was designed to develop a facile and inexpensive method to immobilize enzyme on a microreactor. We report on the procedure for immobilizing enzyme on the internal surface of the microchannel by forming an enzyme-polymer membrane through a cross-linking polymerization method.
A typical procedure for the enzyme-membrane embedded microreactor is as follows. Commercially available polytetrafluoroethylene (PTFE) tubing (500 µm inner diameter and 6 cm length) was used for microreactor preparation. Glutaraldehyde (GA) and paraformaldehyde (PA) were employed as bifunctional cross-linker agents to facilitate enzyme–enzyme covalent binding. The GA and PA in phosphate buffered saline (PBS, pH 8.0) were used at 0.25% (v/v) and 4% (v/v), respectively. The enzyme α-chymotrypsin (specific activity = 1000 units mg−1), one of the enzymes used for protein digestion, was used in this study.5 Recently, this process has been crucial for analysis of unknown protein, as in the case of proteomics studies. The enzyme was dissolved in PBS (pH 8.0) and the final concentration equivalent to 10 mg ml−1 was filtered (pore size 0.2 µm). As shown in Fig. 1a, the enzyme and the aldehyde reagents containing PA and GA solutions were separately loaded into a PTFE tube from a 1 ml syringe equipped with a syringe pump (KD Scientific Inc., New Hope, PA, USA) at different pumping rates (0.75 µl min−1 for cross-linker and 0.5 µl min−1 for enzyme). After a few hours, a water-insoluble enzyme-membrane was formed on the inner wall surface of the PTFE tube (Fig. 1b). In the case of batchwise polymerization using chymotrypsin and aldehyde reagents, the enzyme-polymer product aggregated. When this suspended reaction mixture was passed through a PTFE tube, no membrane was observed on the inner wall as expected. This result indicates that polymerizing in a microfluid enables the enzyme-polymer product to form the membrane state on the inner wall of the tube. As shown in Fig. 1c, when enzymes polymerize at the fluid flow of the center region, the polymerizing products tend to drift toward the direction of the flow due to the greater linear velocity compared to the diffusion velocity of chymotrypsin in the center region (data not shown). In contrast, because flow velocity in the region close to the inner wall decreases markedly, it would be difficult for the product polymerizing in such a region to drift forward in the direction of the flow. In addition, the residence time of the chymotrypsin on the PTFE wall surface would be extended due to its adsorption to the hydrophobic surface.6 These factors would lead to promotion of the enzyme-polymer growth at the inner wall as compared to that in the center region of the microchannel. The obtained membrane-formed tube was rinsed with 1 M Tris-HCl (pH 9.0), which simultaneously quenched active aldehyde groups remaining on the membrane. In addition, in order to reduce the resulting Schiff base, the tube was treated with 50 mM NaBH4 in borate buffer (pH 9.0), and then well washed with PBS. The microreactor preparation was performed at 4 °C. The microreactor tube was filled with PBS and stored at 4 °C. Even with storage of the microreactor in a dry environment, the protease activity was recovered by re-swelling of the dry membrane with a buffer. In addition, there was no change in the mechanical strength of the membrane. In order to estimate the amount of the immobilized enzyme, the tube was washed with distilled water, and then the enzyme-membrane was dried by a vacuum pump (Fig. 1b), and peeled from the tube with forceps. The obtained membrane was hydrolyzed and the protein content was quantified by amino acid analysis. The amount of protein content in the membrane was 380 µg in a microreactor. Calculating from the size of α-chymotrypsin, up to 15.2 nmol of enzyme molecule could be immobilized in the membrane on the inner wall of the microreactor. In order to examine the loss of enzyme molecule from the membrane during the enzyme reaction, 50 ml of distilled water was flushed through the microreactor at a flow rate of 10 µl min−1 for about 3.5 days, and the collected and concentrated sample was estimated by amino acid analysis. The amount of protein that leaked out of the enzyme-membrane into the flowing water was a few micrograms. Furthermore, the microreactor was pressurized by a rapid flow of water at a flow rate of 10 ml min−1 (the corresponding pressure loss was 6.52 kg cm−2). Destruction of the membrane and membrane detachment from the microreactor were not observed. These results demonstrate that the enzyme-membrane covalently captures enzymes and has sufficient mechanical strength for microfluidic system applications. We applied this method to fused-silica or PEEK tubes. Enzyme-immobilized membrane was formed in all cases. Thus, it can be said that this method is universally applicable.
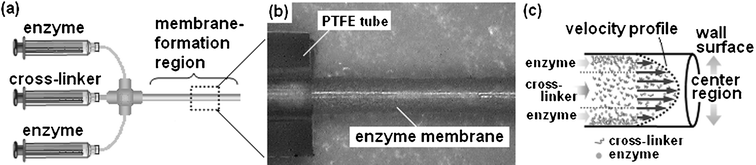 | ||
| Fig. 1 Preparation of enzyme-membrane on the inner wall of a PTFE tube. a) Enzyme and aldehyde solutions were each charged into a 1 ml syringe, the solutions were supplied to a PTFE tube using a syringe pump. b) Cylindrical enzyme-membrane (dry state) exposed from PTFE tube, which forms on the inner wall of the tube. c) Possible mechanism of polymerization process of enzyme and cross-linker reagent in a microchannel. | ||
We examined the reaction efficiency using this enzyme-immobilized microreactor. Hydrolysis of N-glutaryl-L-phenylalanine p-nitroanilide (HOOC(CH2)3CONHCH(CH2C6H5)CONHC6H4NO2; GPNA)
(1 mM in PBS, pH 7.5), a synthetic substrate for α-chymotrypsin, was used for evaluation. The reaction was performed in a thermostated incubator controlled at 37 °C. In the microreactor, the hydrolysis reaction was completed at a flow rate of 4 µl min−1, which yields a residence time of 3 min (Fig. 2). The kinetic constants Km and Vmax calculated from a Lineweaver–Burke plot were 0.50 mM and 0.77 mM min−1, respectively. There was no significant difference in the hydrolysis efficiency compared to that of solution-phase batchwise reaction using the same enzyme (3 mg ml−1)/substrate molar ratio, which took 170 s to complete the reaction. However, no membrane formation was observed in microreactors prepared by using GA (0.25%) only or PA (4%) only as cross-linkers (captured proteins = 30 µg and below) and these reactors showed relatively lower protease activity (Fig. 2). Using the concentrated PA (16%) also led to a comparable result. The mechanical strength of the obtained membrane was relatively low, and it also captured a small amount (about 30 µg and below) of the protein. In contrast, although the concentrated GA (2%) only enabled strong membrane-formation and high protease activity by the strict control of the reaction time, it was very difficult to control the enzyme-polymerization in a micro-space due to rapid aggregation of enzymes. This procedure frequently led to pronounced evenness of the membrane and tube obstruction, resulting in very low reproducibility in the enzyme immobilization. Also, enzymes might be deactivated by over-reactivity of GA against the protein.† PA is composed mainly of formaldehyde and its self-polymer in the buffer. Its cross-linking reaction against protein is well known to be very complex and the chemical stability of the cross-linked polymer would be lower than that of GA with stable aldehyde-polymer including an α,β unsaturated structure (![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) [C(CHO)–CH2CH2–CH]n
[C(CHO)–CH2CH2–CH]n![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ).7 Thus, the membrane preparation using PA only would be unsuccessful as compared to that using GA. However, addition of PA in rapid enzyme-polymerization by GA could provide an appropriate and controllable rate of the polymerization for enzyme-membrane formation. The combination of GA and PA was a key factor in the preparation of the microreactor. Furthermore, it was found that the combination ratio of GA/PA and/or the total amounts of aldehyde reagents were also critical because membranes could not be formed successfully in preparations using other GA/PA ratios and/or using other aldehyde amounts.†
).7 Thus, the membrane preparation using PA only would be unsuccessful as compared to that using GA. However, addition of PA in rapid enzyme-polymerization by GA could provide an appropriate and controllable rate of the polymerization for enzyme-membrane formation. The combination of GA and PA was a key factor in the preparation of the microreactor. Furthermore, it was found that the combination ratio of GA/PA and/or the total amounts of aldehyde reagents were also critical because membranes could not be formed successfully in preparations using other GA/PA ratios and/or using other aldehyde amounts.†
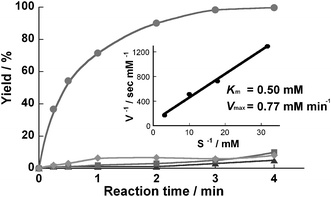 | ||
| Fig. 2 Hydrolysis of GPNA in a microreactor. A 1 mM solution of substrate in Tris buffer was charged into a microreactor which was prepared using both PA and GA (●), GA only (▲), and PA only (■) as cross-linker, at 37 °C. Product was analyzed by RP-HPLC. When the microreactor prepared normally was treated with PBS solution containing 0.1 mM phenylmethanesulfonyl fluoride which irreversibly inhibits chymotrypsin, the protease activity of the microreactor decreased markedly (♦). A Lineweaver–Burke plot was built from the kinetic data of the microreactor prepared using both PA and GA. | ||
High stability of the enzymatic activity for protein digestion should be an important feature of the enzyme-reactors used in proteomics studies because digestion is often carried out under denaturating conditions to achieve an efficient and rapid digestion of protein substrates.8 The present microreactor was durable for at least 40 days while the proteolytic activity of the free chymotrypsin solution became progressively lower (Fig. 3a). In addition, this microreactor could maintain the hydrolysis yield at 90% and above in a continuous flow (4 µl min−1) of substrate solution for a few days (data not shown). Also, the microreactor showed resistance to a chemical denaturant like urea and an organic solvent like dimethyl sulfoxide (DMSO) as compared to free chymotrypsin (Fig. 3b). This microreactor demonstrated high stability and utility for hydrolysis under denaturation conditions. Our data were consistent with previous reports using a GA cross-linking method which provided enzymes with high stability under denaturation conditions.9 This effect was considered to result from the rigid conformation and avoidance of autolysis of the immobilized enzyme.10 An application of an enzyme-immobilizing microreactor for protein digestion was examined using known protein substrates. This reaction was performed at 70 µg ml−1 myoglobin and 50 µg ml−1 insulin solution at 37 °C affording a residence time of 10 min to give digestion yields of 70% and 59%, respectively (data not shown). In comparison, the digestion of the same proteins by free protease according to the conventional procedure11 (enzyme–substrate protein mass ratio 1 ∶ 50) took several hours to reach the same digestion yield (data not shown). These results showed the high potential of this microreactor for enzymatic digestions. In addition, because PTFE tubing is used as standard equipment on various analytical instruments such as liquid chromatographs and mass spectrometers, the application of this microreactor to on-line analysis systems is considered to be not technically difficult.
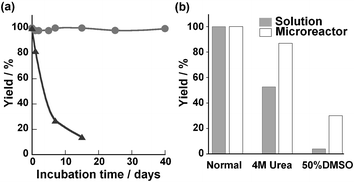 | ||
| Fig. 3 Stability of the immobilized enzyme in the microreactor and of free chymotrypsin solution (3 mg ml−1). a) Durability of enzyme catalysts. The graph shows comparison of reaction yield of microreactor (●) and the solution-phase reaction (▲). The microreactor or enzyme solutions were incubated for various times at 37 °C, and then their protease activities against 1 mM GPNA were measured. b) Stability of enzymes towards denaturing agents (4 M urea and 50% DMSO), the protease activities of the microreactor were compared with those of free-enzyme solution. Each reaction time was 3 min in the microreactor and 20 min in batchwise solution, which was required for completion of the reaction. | ||
Recent proteomic investigations have been directed toward high-throughput and parallel analyses of various unknown proteins. A microreactor would be suited to such an analysis system. However, one of the frequent problems encountered when using a microreactor is non-specific adsorption of protein. Therefore, the microreactor should be disposable and cost as well as efforts for preparation should be reduced. In a previous report, we developed an enzyme-immobilized glass-capillary microreactor without technical difficulties, which could achieve efficient enzymatic reactions. However, the preparation method requires time, effort, and large amounts of reagents for every step. In our present study, we developed a facile method to inexpensively prepare the enzyme-immobilized microreactor by formation of the enzyme-polymer membrane. The preparation cost of the present microreactor is nearly one-tenth of the previously developed microreactor. This preparation method also has promising applications to other enzymes. Actually, we have successfully achieved the preparation of a trypsin-embedded microreactor (data not shown). Trypsin is an enzyme frequently used in proteomics studies. In the case of trypsin (specific activity = 205 units mg−1), the concentration (< 1 µg µl−1) sufficient to form a membrane with high protease activity was significantly lower than that of α-chymotrypsin. Enzyme concentrations used in this preparation method seem to be different for different enzymes. This might be caused by the differences in purity of the enzymes, and the number and location of amino groups on the enzyme surface.
In summary, we have developed an enzyme-immobilizing microreactor by cross-linking polymerization. This method may also be of interest in various micro-analysis systems involving immobilized biomolecules such as enzymes, antibodies, glycoproteins, and other functional proteins.
Notes and references
- (a) V. Hessel, S. Hardt and H. Lowe, Chemical Micro Process Engineering, Wiley-VCH, Weinheim, 2004 Search PubMed; (b) P. D. I. Fletcher, S. J. Haswell, E. Pombo-Villar, B. H. Warrington, P. Watts, S. Y. F. Wong and X. Zhang, Tetrahedron, 2002, 58, 4735 CrossRef CAS; (c) T. Chovan and A. Guttman, Trends Biotechnol., 2002, 20, 116 CrossRef CAS; (d) J. Khandurina and A. Guttman, J. Chromatogr., 2002, 943, 159 CrossRef CAS.
- (a) K. Kusakabe, D. Miyagawa, Y. Gu, H. Maeda and S. Morooka, J. Chem. Eng. Jpn., 2001, 34, 441 Search PubMed; (b) C. de Bellefon, N. Pestre, T. Lamouille, P. Grenouillet and V. Hessel, Adv. Synth. Catal., 2003, 345, 190 CrossRef CAS; (c) X. Li, H. Wang, K. Inoue, M. Uehara, H. Nakamura, M. Miyazaki, E. Abe and H. Maeda, Chem. Commun., 2003, 964 RSC.
- N. Lion, F. Reymond, H. H. Giraut and J. S. Rossier, Curr. Opin. Biotechnol., 2004, 15, 31 CrossRef CAS.
- (a) M. Miyazaki, J. Kaneno, M. Uehara, H. Fujii, M. Shimizu and H. Maeda, Chem. Commun., 2003, 648 RSC; (b) J. Kaneno, R. Kohama, M. Miyazaki, M. Uehara, K. Kanno, M. Fujii, H. Shimizu and H. Maeda, New J. Chem., 2003, 27, 1265 Search PubMed; (c) M. Miyazaki, J. Kaneno, R. Kohama, K. Kanno, M. Fujii, H. Shimizu and H. Maeda, Chem. Eng. J., 2004, 101, 277 CrossRef CAS; (d) M. Miyazaki, J. Kaneno, S. Yamaori, T. Honda, M. P. P. Briones, M. Uehara, K. Arima, K. Kanno, K. Yamasita, Y. Yamaguchi, H. Nakamura, H. Yonezawa, M. Fujii and H. Maeda, Protein Pept. Lett., 2005, 12, 207–210 Search PubMed.
- D. W. Cleveland, S. G. Fischer, M. W. Kirschner and U. K. Laemmli, J. Biol. Chem., 1977, 252, 1102 CAS.
- (a) M. Morra and C. Cassinelli, J. Biomed. Mater. Res., 1995, 29, 39 CrossRef CAS; (b) S. Noinville, M. Revault and M.-H. Baron, Biopolymers, 2002, 67, 323 CrossRef CAS.
- (a) R. E. Feeney, G. Blankenhorn and H. B. F. Dixon, Adv. Protein Chem., 1975, 29, 136; (b) P. Monsan, G. Puzo and H. Mazarguil, Biochimie, 1975, 57, 1281 CAS; (c) Y. Kitamoto and H. Maeda, J. Biochem., 1980, 87, 1519 CAS.
- A. K. Palm and M. V. Novotny, Rapid Commun. Mass Spectrom., 2004, 18, 1374 CrossRef CAS.
- N. L. S. Clair and M. A. Navia, J. Am. Chem. Soc., 1992, 114, 7314 CrossRef CAS.
- M. N. Gupta and B. Mattiasson, in Methods of Biochemical Analysis: Bioanalytical Applications of Enzymes, vol. 15. John Wiley, New Jersey, pp. 1–34 Search PubMed.
- D. S. Peterson, T. Rohr, F. Svec and J. M. J. Frechet, J. Proteome Res., 2002, 1, 563 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: experimental results on reproducibility, protease activity per enzyme, and influences of the GA/PA ratio and the concentration of total aldehyde in microreactor preparation. See http://dx.doi.org/10.1039/b510605b |
| This journal is © The Royal Society of Chemistry 2005 |
