Fluorescence resonance energy transfer between a quantum dot donor and a dye acceptor attached to DNA†
Dejian
Zhou
*ab,
Joe D.
Piper
b,
Chris
Abell
b,
David
Klenerman
b,
Dae-Joon
Kang
*ac and
Liming
Ying
*b
aNanoscience Centre, University of Cambridge, 11 J J Thomson Avenue, Cambridge, UK CB3 0FF. E-mail: dz209@cam.ac.uk; Fax: +44 1223 760309; Tel: +44 1223 760312
bDepartment of Chemistry, University of Cambridge, Lensfield Road, Cambridge, UK CB2 1EW. E-mail: ly206@cam.ac.uk; Fax: +44 1223 336362; Tel: +44 1223 336421
cSungkyunkwan Advanced Institute of Nanotechnology and Department of Physics, Sungkyunkwan University, Suwon, 440-746, Korea. E-mail: djk1003@cam.ac.uk; Fax: +44 1223 760309; Tel: +44 1223 760311
First published on 6th September 2005
Abstract
We show that direct coupling of a dye-labelled DNA (acceptor) to a quantum dot (QD) donor significantly reduces the donor–acceptor distance and improves the FRET efficiency: a highly efficient FRET (∼88%) at a low acceptor-to-donor ratio of 2 has been achieved at the single-molecule level.
Quantum dots (QDs) are of great interest currently due to their unique size-dependent, symmetric, narrow and stable emissions, allowing for prolonged observation and multiplexing.1 They are excellent donors in fluorescence resonance energy transfer (FRET) based applications due to their narrow emission and broad excitation spectra, enabling the effective separation of the donor and acceptor fluorescence, and the selection of a wide range of excitation wavelengths to reduce background.2–4 Studies on the FRET between two different coloured QDs,2 QD and dye-labelled biomolecule,3–5 QD and gold nanoparticle,6 and QD and dye-labelled polymer7 have been reported. Most of these studies have been carried out in bulk solutions,2–4 providing only the ensemble averaged FRET information. Further, they were mostly based on QD and dye-labelled proteins, which led to low FRET efficiencies due to the large size of the QD and protein.3–5 Recently, single-molecule FRET between a protein-coated QD and dye-labelled DNA has also been reported, but the FRET efficiency is low (i.e. 1–2%).5 To achieve high FRET, a number of acceptors (>10) are required for each QD in the QD–protein design.3,4 This significantly reduces the sensitivity of these conjugates as FRET-based sensors, since a number of binding events to the conjugated proteins are required to produce a detectable change in FRET signal. In this communication, we directly couple a dye-labelled DNA acceptor to a QD donor through a thiol linker. This reduces the donor–acceptor distance, and thus significantly increases the FRET efficiency. The FRET between the QD and dye-labelled DNA was studied in both bulk solution and at the single-molecule level.
Our design of QD–DNA conjugate is shown schematically in Fig. 1. The acceptor, an Alexa 594 fluorophore labelled double-stranded (ds) DNA,‡ is directly coupled to the donor, a CdSe/ZnS core-shell QD, through a C6-thiol linker.8 Trioctyl-phosphine oxide (TOPO) capped CdSe/ZnS core-shell QD (peak emission ∼553 nm, from Evident Technologies, New York) was treated with 3-mercaptopropionic acid (MPA) in a mixed solvent of chloroform/methanol at pH ∼10 to produce a water-soluble MPA-capped QD (MPA–QD) following a literature procedure.9 The MPA–QD shows the same narrow emission (FWHM 30 nm, quantum yield ∼15%) with the peak slightly red-shifted to 558 nm (Fig. 2). The QD emission overlaps the absorption spectrum of the dye, suggesting that efficient FRET between the QD donor and dye acceptor can take place. Based on the spectral overlap, the bulk quantum yield of the QD and the absorption coefficient of the dye, we estimate a Förster distance R0 (a distance showing 50% FRET efficiency) of ∼4.2 nm in our FRET system.4 There is little overlap between the donor and acceptor fluorescence (Fig. 2), allowing for the effective separation of donor fluorescence from that of the acceptor. The Alexa 594-labelled dsDNA (at different DNA to QD ratios) was conjugated to the MPA–QD following a literature procedure.8
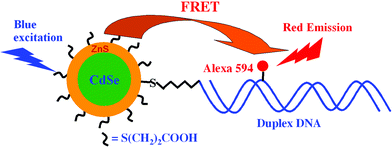 | ||
| Fig. 1 Schematic of the QD-dye-labelled DNA FRET system. The acceptor, an Alexa 594-labelled duplex DNA, is conjugated to the donor, a CdSe/ZnS core/shell QD, through a C6-thiol linker. This reduces the donor–acceptor distance and significantly increases the FRET efficiency. | ||
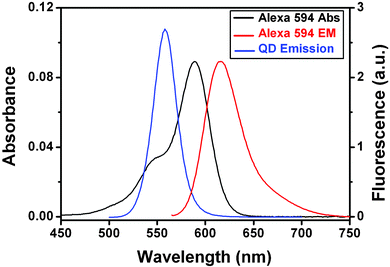 | ||
| Fig. 2 Absorption spectrum (black line, left scale) and fluorescence spectrum (red line, right scale) of Alexa 594 labelled DNA (1 µM), and fluorescence spectrum of the MPA-capped CdSe/ZnS core/shell QD (blue line, right scale). All measurements were carried out in Tris buffer (10 mM Tris·HCl, 100 mM NaCl, pH 7.6) at room temperature. | ||
To achieve a high conjugation (FRET) efficiency between the QD and the dye-labelled DNA, it was found necessary to remove the excessive unbound MPA used to render the QD water-soluble. This was achieved by precipitation with ethanol followed by centrifugation at 14000 rpm for 20 min. Unbound MPA is soluble in ethanol but the MPA–QD is not under centrifugation conditions. Removal of the excess MPA eliminated the competition for QD binding from MPA, and greatly increased the conjugation efficiency of the DNA to the QD. Fluorescence spectra of the QD–DNA conjugates at different ratios of DNA to QD are shown in Fig. 3a. Conjugation of the dye-labelled DNA to the QD quenches the donor (QD) fluorescence at 560 nm, while at the same time enhancing the acceptor (dye) fluorescence at 618 nm through FRET. The estimated FRET efficiency, defined as IA/(IA + ID) where IA and ID are the acceptor and donor intensities, increases with the ratio of DNA to QD initially, but quickly levels off at a DNA : QD ratio of 0.5, where an ∼50% FRET efficiency is obtained (Fig. 3b). The FRET efficiency is higher than anticipated from the equation, E = nR06/(nR06 + r6), for FRET with n identical acceptors interacting with a single donor.4 By using the FRET efficiencies and the equation above, we estimate the average donor–acceptor distance r as 4.10, 3.80, 4.33 and 4.60 nm for DNA : QD ratios of 0.25, 0.50, 1.0 and 2.0, respectively. The predicted distance from dye to the QD center is 4.60 nm, taking into account the size of the QD and assuming the DNA is extended. Thus the experimental value matches the prediction only at the DNA : QD ratio of 2, and at all other ratios, the distance obtained is markedly smaller than predicted.
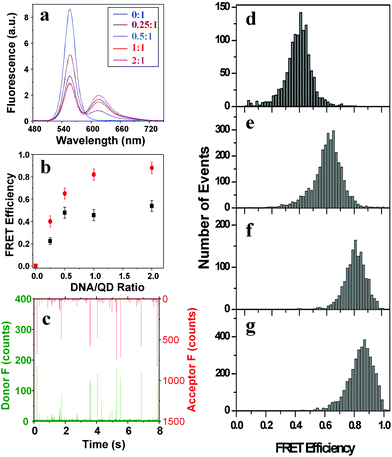 | ||
| Fig. 3 (a) Fluorescence spectra (excited at 450 nm) of the DNA–QD conjugates at different DNA/QD ratios prepared after removal of the free MPA. (b) Plot of the apparent FRET efficiency (IA/(IA + ID)) versus the DNA/QD ratios from the bulk (black squares) and single-molecule studies (red dots). (c) Representative fluorescence burst trajectories of the QD–DNA conjugates at 1 : 1, the green bursts are from the donor (QD) and the red bursts are from the acceptor (Alexa 594). (d–g) FRET histograms of the QD–DNA conjugates at different DNA to QD ratios, (d) 0.25; (e) 0.50; (f) 1.0, and (g) 2.0. | ||
A single-molecule study, that simultaneously records the QD (donor) and dye (acceptor) fluorescence bursts as each DNA–QD conjugate diffuses through the laser focus (probe volume),10 revealed very bright acceptor fluorescence bursts (up to several hundred counts per ms) at low DNA to QD ratios (Fig. 3c). They are much brighter than the typical bursts from a single fluorophore (∼10 counts per ms) under identical conditions. The acceptor bursts at DNA : QD ratio ≤1 are stronger than those at higher ratios (i.e. 2), contradicting the idea that a higher DNA : QD ratio should lead to more intense acceptor bursts. It was also found necessary to agitate the solution with a pipette during the collection of fluorescence bursts at the DNA : QD ratio of ≤1, but this was not necessary at higher ratios (i.e. 2.0). This suggests that the QD–DNA conjugates are aggregated at the low ratios (i.e. ≤1), but not or only slightly aggregated at the higher ratios. To verify this, we immobilised the QD–DNA conjugates on a flat template stripped gold surface (TSG, typical roughness <0.2 nm) modified with a self-assembled monolayer of 6-mercapto-N-hexylpyridinium bromide, a positively charged thiol.11 Tapping mode AFM topographic images12 showed that at a DNA : QD ratio of 0.25, the QD–DNA conjugates were heavily aggregated (ESI, Fig. S1a†), but they were mostly isolated at a DNA : QD ratio of 2 (ESI, Fig. S1b), suggesting that the single-molecule FRET measured at the DNA : QD ratio of 2 is indeed from an individual single QD–DNA conjugate.
Fig. 3c–g show FRET histograms at different DNA : QD ratios. All the histograms show a single distribution of FRET efficiencies, with the mean FRET increasing from 0.40 to 0.88 as the DNA : QD ratio increases from 0.25 to 2.0 (Fig. 3b). The FRET efficiency obtained from the single-molecule study is higher than that of the bulk, presumably because the single molecule method only detects bright QD–DNA conjugates (a threshold of 50 counts per ms bin for the sum of the donor and acceptor fluorescence signals was used to differentiate single molecule bursts from the background), while the bulk method measures signals from all QD–DNA conjugates, bright and dim. If we assume a quantum yield of 0.9 for the bright QDs, we would get a Förster distance around 5.6 nm and then the FRET efficiency would be 0.77 for the 1 : 1 DNA/QD conjugate, this is quite consistent with our single-molecule result shown in Fig. 3f. Thus we have shown that it is possible to achieve a high FRET efficiency between the QD and dye-labelled DNA at the single-molecule (QD) level at a DNA : QD ratio of 2.0 with little aggregation.
The aggregation of the QD–DNA conjugates observed at the low DNA : QD ratios may be due to the interaction between the QD and the DNA backbone, and may be a common feature for QD–biomolecule conjugates that have been used in the FRET studies.2–4 However, without an independent analysis by the single-molecule method, this information would have been buried by the ensemble-average provided by the bulk methods. Thus the single-molecule method provides a facile way for checking the sample aggregation status and guiding the optimizing of sample preparations.
It should be noted that the removal of the excess MPA and controlling the DNA : QD ratios are key to achieve high FRET without significant aggregation. Conjugation of the DNA to the MPA–QD prior to the removal of the excess unbound MPA only produced a low FRET efficiency of around 2.6% per acceptor, where the apparent overall FRET increases linearly with the DNA : QD ratio (ESI, Fig. S2†), presumably because not all DNAs have conjugated to the QD due to the competition for QD binding from the unbound MPA. The single-molecule study revealed a broad distribution of the FRET efficiencies (ESI, Fig. S2c†), reflecting the heterogeneity of the sample because a variable number of the labelled DNAs may attach to a single QD.5 After removal of the excess unbound MPA, the use of a higher DNA : QD ratio reduces the QD–DNA conjugate aggregation, but this was found to have a limit. When the DNA : QD ratio (unlabelled, same sequence) is further increased to 4, the QD fluorescence is greatly reduced and accompanied by significant surface defect emission (ESI, Fig. S3†), presumably because the QD fluorescence is very sensitive to the environment and its surface ligand coating.13
In summary, we have shown that direct coupling of the dye-labelled thiolated DNA (acceptor) to the QD (donor) reduced the donor–acceptor distance, leading to a greatly improved FRET efficiency. Unlike the QD–protein conjugates, where a number of acceptors are needed for each QD to achieve moderate FRET, we show that highly efficient FRET (∼88%) can be obtained at low acceptor to donor ratios (i.e. 2) at the single-molecule level with little aggregation. This may prove to be particularly useful in making sensitive FRET-based sensors, where a single interaction with the acceptor can produce a big change in FRET signal. These QD–DNA conjugates may also prove to be useful building materials for the assembly of novel functional nanostructures,14 and work on these aspects is currently underway.
We thank the Interdisciplinary Research Collaboration in Nanotechnology (Cambridge Nano IRC) and the Biotechnology and Biological Sciences Research Council (BBSRC) for funding this project.
Notes and references
- P. Alivisatos, Nat. Biotechnol., 2004, 22, 47 CrossRef; X. Michalet, F. F. Pinaud, L. A. Bentolila, J. M. Tsay, S. Doose, J. J. Li, G. Sundaresan, A. W. Wu, S. S. Gambhir and S. Weiss, Science, 2005, 307, 538 CrossRef CAS; M. Han, X. Gao, J. Z. Su and S. Nie, Nat. Biotechnol., 2001, 19, 631 CrossRef CAS.
- R. Wargnier, A. V. Baranov, V. G. Maslov, V. Stsiapura, M. Artemyev, M. Pluot, A. Sukhanova and I. Nabiev, Nano Lett., 2004, 4, 451 CrossRef CAS.
- N. N. Mamedova, N. A. Kotov, A. L. Rogach and J. Studer, Nano Lett., 2001, 1, 281 CrossRef CAS; A. R. Clapp, I. L. Medintz, J. M. Mauro, B. R. Fisher, M. G. Bawendi and H. Mattoussi, J. Am. Chem. Soc., 2004, 126, 301 CrossRef CAS; I. L. Medintz, S. A. Trammell, H. Mattoussi and J. M. Mauro, J. Am. Chem. Soc., 2004, 126, 30 CrossRef CAS.
- I. L. Medintz, A. R. Clapp, H. Mattoussi, E. R. Goldman, B. Fisher and J. M. Mauro, Nat. Mater., 2003, 2, 630 CrossRef CAS; E. R. Goldman, I. L. Medintz, J. L. Whitley, A. Hayhurst, A. R. Clapp, H. T. Uyeda, J. R. Deschamps, M. E. Lassman and H. Mattoussi, J. Am. Chem. Soc., 2005, 127, 6744 CrossRef CAS; E. Oh, M. Y. Hong, D. Lee, S. H. Nam, H. C. Yoon and H. S. Kim, J. Am. Chem. Soc., 2005, 127, 3270 CrossRef CAS.
- S. Hohng and T. Ha, ChemPhysChem, 2005, 6, 956 CrossRef CAS.
- Z. Gueroui and A. Libchaber, Phys. Rev. Lett., 2004, 93, 166108 CrossRef; L. Dyadyusha, H. Yin, S. Jaiswal, T. Brown, J. J. Baumberg, F. P. Booy and T. Melvin, Chem. Commun., 2005, 3201 RSC.
- I. Potapova, R. Mruk, C. Hübner, R. Zentel, T. Basché and A. Mews, Angew. Chem., Int. Ed., 2005, 44, 2437 CrossRef CAS.
- G. P. Mitchell, C. A. Mirkin and R. L. Letsinger, J. Am. Chem. Soc., 1999, 121, 8122 CrossRef CAS.
- S. F. Wuister, I. Swart, F. van Driel, S. G. Hickey and C. Donega, Nano Lett., 2003, 3, 503 CrossRef CAS; D. J. Zhou, A. Bruckbauer, C. Abell, D. Klenerman and D.-J. Kang, Adv. Mater., 2005, 17, 1243 CrossRef CAS.
- L. M. Ying, J. J. Green, H. T. Li, D. Klenerman and S. Balasubramanian, Proc. Natl. Acda. Sci. U. S. A., 2003, 100, 14629 Search PubMed.
- D. J. Zhou, X. Z. Wang, L. Birch, T. Rayment and C. Abell, Langmuir, 2003, 19, 10557 CrossRef CAS.
- D. Zhou, A. Bruckbauer, L. M. Ying, C. Abell and D. Klenerman, Nano Lett., 2003, 3, 1510.
- Y. Wang, Z. Y. Tang, M. A. Correa-Duarte, L. M. Liz-Marzan and N. A. Kotov, J. Am. Chem. Soc., 2003, 125, 2830 CrossRef CAS; W. Guo, J. J. Li, Y. A. Wang and X. Peng, J. Am. Chem. Soc., 2003, 125, 3901 CrossRef CAS.
- A. Fu, C. M. Micheel, J. Cha, H. Chang, H. Yang and A. P. Alivisatos, J. Am. Chem. Soc., 2004, 126, 10832 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Details of the sample preparation, experimental procedures, and AFM topographic images showing the aggregation of the QD–DNA conjugates. See http://dx.doi.org/10.1039/b508911e |
| ‡ The sequence of the duplex DNA is as follows: 5′-CAT AAA AGA GCT CCA TAT CCA ACC TGC ACG-3′ 3′-GTA TTT TCT CGA GGT ATA GGT TGG ACG TGC-5′ where the base T shown in bold is labelled with an Alexa 594 fluorophore. The DNA is likely to attach to the QD first through its thiol linker since this interaction is strong and fast (ligand exchange between a TOPO capped QD and a thiol is complete in minutes).8 The DNA phosphate backbone and base unit can also bind to the Zn2+ ions on the QD surface, but this process is much slower (i.e. several days). When the free MPA is removed, direct binding between the DNA and other QDs becomes possible and causes aggregation. |
| This journal is © The Royal Society of Chemistry 2005 |
