Identification of broad specificity P450CAM variants by primary screening against indole as substrate
Ayhan
Çelik
,
Robert E.
Speight
and
Nicholas J.
Turner
*
School of Chemistry, University of Edinburgh, King's Buildings, West Mains Road, Edinburgh, UK EH9 3JJ. E-mail: n.j.turner@ed.ac.uk; Fax: +44 131 6504719
First published on 5th July 2005
Abstract
High-throughput screening of cytochrome P450CAM libraries, for their ability to oxidise indole to indigo and indirubin, has resulted in the identification of variants with activity towards the structurally unrelated substrate diphenylmethane.
Cytochrome P450 monooxygenases (P450s) are a diverse superfamily of enzymes present in plants, animals and microorganisms. The function of these enzymes varies depending on the host organisms. For example, in plants they are involved in both primary and secondary metabolism. In mammals, P450s play a central role in detoxification pathways. Bacterial P450s are involved in the degradation of various natural substrates. The most extensively studied P450 monooxygenase is cytochrome P450CAM from Pseudomonas putida. This enzyme, which catalyses the stereoselective hydroxylation of camphor to 5-exo-hydroxy camphor,1 serves as a model system for studying the mechanism of other P450 monooxygenases. Typically (type I) three separate proteins are required for functional activity, namely a cytochrome P450 reductase (putidadoxin reductase), an iron–sulfur electron transfer protein (putidaredoxin) and the cytochrome P450 monooxygenase enzyme. Thus, unlike cytochrome P450BM3 from Bacillus megaterium, which contains both a haem-containing P450 domain and a cytochrome P450 reductase domain in a fused form, P450CAM is not a catalytically self sufficient enzyme. For applications in the synthesis of fine chemicals, bioremediation and the development of herbicide resistant plants, an efficient enzyme system is clearly desirable. For P450CAM the three proteins have been engineered into a fused system.2 However, in comparison to the wild type system, the activity was found to be quite low. Moreover, the lack of effective high throughput screening methods has limited the number of variants that can be screened in order to identify modified enzymes with altered properties (e.g. broadened substrate specificity or enhanced rate of electron transfer between P450CAM and putidaredoxin).
Recently we reported an approach for identifying P450CAM variants based upon generating fully randomised active site libraries coupled with GC-MS screening for product formation.3 The use of GC-MS allows only ca. 500 variants per day to be screened and hence places limitations on the size of libraries that can be evaluated. Hence we sought to develop an alternative approach that would be able to handle much larger libraries.
The basis for this method is that a library of P450CAM mutants, in which the active site residues of Tyr96 and Phe98 were combinatorially randomised, was screened for the ability to catalyse hydroxylation of indole 1 to 3-hydroxy indole 2, which subsequently undergoes spontaneous air oxidation to produce the insoluble dye indigo 3 (Scheme 1). This dye can easily be detected at very low levels and can be used as an indication of enzyme activity either on LB-agar or liquid media. During the screening of P450CAM mutant enzymes, which were functionally co-expressed with their natural redox partners in E. coli for activities towards various substrates (e.g. diphenylmethane), we observed that some of the mutant enzymes produced a dark blue pigment within the biotransformation wells. Although similar observations of this kind have been reported previously for several mono- and di-oxygenase enzymes, including styrene monooxygenase,4 naphthalene dioxygenase,5 cytochrome P450s2A6 and 2E16 and P450BM3,7 there are no previous reports of the involvement of P450CAM in the pigment formation. We subsequently identified the pigments formed in bacterial cultures containing a functional recombinant P450CAM system and examined the ability of different P450CAM variants to catalyze indigo formation.
 | ||
| Scheme 1 | ||
A library of P450CAM mutants8 (Tyr96/Phe98) was screened for hydroxylation of various substrates in a 96-well microtitre plate format (only 48 wells were used), in which each well contained a mutant of P450CAM (at least one amino acid mutation). Generation of the blue pigment was observed in several wells (Fig. 1, Panel A). Addition of exogenous indole (1 mM) resulted in enhanced colour formation with the same variants (Fig. 1, Panel B). Pigment formation in the absence of added indole is not surprising, since it was previously demonstrated that a tryptophanase in E. coli converts tryptophan to indole.9 Wild type P450CAM does not form the pigment regardless of the presence or absence of indole in the growth medium. Therefore colour formation is clearly associated with the variant enzymes. Small scale biotransformations were carried out using mutant 43.10 The pigment produced in the P450CAM mutant culture was separated into four components. The visible spectra of two major components give absorbance at max, 602, 550 nm, which correspond to indigo 3 and indirubin 4.6,7 Positive ESI mass spectrometry yielded an apparent MH+ ion at m/z 263 as expected for 3 and 4. The yield of indigo formed using mutant 43 was approximately 9 mg/L, which is comparable to that obtained with other monooxygenases.5
 | ||
| Fig. 1 Microtitre plate assay showing indigo formation in absence (A) and presence (B) of indole by P450CAM mutants. | ||
We then compared the variants showing activity towards indole with a structurally different substrate, diphenylmethane. Fig. 2 presents the relative activities towards indole and diphenylmethane, the latter determined by GC-MS. Interestingly the ability of the variants to hydroxylate indole correlates very well with their ability to hydroxylate diphenylmethane (ca. 96% correlation). Only two of the 48 mutants (17 and 45) which displayed activity towards diphenylmethane showed little or no activity for indole.
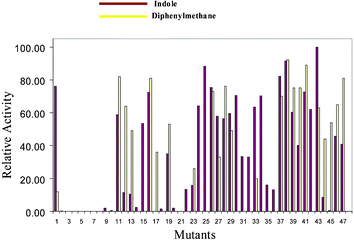 | ||
| Fig. 2 Comparison of the relative activities of specific variants of P450CAM using indole (oxidation to indigo) and diphenylmethane (hydroxylation) as substrates. | ||
The structure–function relationship of the variant enzymes can be summarised as follows. Maintaining Phe98 and mutating Tyr96 to Ala, Cys, Gln, Gly, Met, Ser and Thr resulted in good activity towards indole. The order of activities was found to be as follows: Met > Ala > Cys > Thr = Gln = Ser > Gly. Sulfur-containing amino acids (Met, Cys) showed amongst the highest activities.
These results may explain why wild type P450CAM is unable to hydroxylate indole. Unlike camphor, indole lacks the carbonyl group that interacts with the hydroxyl group of Tyr96. Furthermore, mutation of Phe98 to Leu, and Tyr96 to Ala, Gly, Met, Ser and Thr provided additional confirmation of the adverse effect of hydroxyl-containing amino acids in the active site of P450CAM for hydroxylation of indole type substrates. This is probably due to interference with hydrogen bonding. Mutation of Tyr96 to Gly and Phe98 to Leu, Phe and Trp showed activities in the following order: Leu > Trp > Phe. This result emphasises the importance of maintaining a significant degree of hydrophobicity in the active site. Additionally, since indole is sterically less demanding than camphor, there is no restriction for indole binding and hydroxylation in the presence of these aliphatic and aromatic amino acids. Effects of further mutations of this kind (Phe98 and Tyr96) to various amino acids are summarised in Table 1. In short, amino acid substitutions identified in this study promote binding and oxidation of unnatural substrates.
| 96 | 98 | Relative activitya | ||
|---|---|---|---|---|
| a Absorbance at 602 nm due to indigo formation was used as a measure of relative activities for the mutants. | ||||
| Wild-Type | 0 | Tyr | Phe | 0 |
| Mutants | 1 | Gly | Leu | 76 |
| 12 | Phe | Gly | 12 | |
| 17 | Ser | Glu | 0 | |
| 19 | — | Met | 35 | |
| 23 | Phe | — | 15 | |
| 24 | Met | Leu | 64 | |
| 27 | Gln | Phe | 58 | |
| 28 | Ser | Phe | 56 | |
| 29 | Ala | Leu | 59 | |
| 38 | Ala | Phe | 91 | |
| 39 | Thr | Phe | 60 | |
| 40 | Ser | Leu | 40 | |
| 41 | Cys | Phe | 72 | |
| 43 | Met | Phe | 100 | |
| 44 | Gly | Phe | 8 | |
| 45 | Ser | His | 0 | |
| 46 | Thr | Leu | 46 | |
| 47 | Gly | Trp | 41 | |
Such high correlation suggested that it might be possible to pre-screen large libraries using indigo production as an indicator. Accordingly, the entire library of P450CAM Y96/F98 variants (400 possible variants) was screened in this way resulting in the selection of 96 (out of 980) colonies that appeared to be blue, a hit rate of approximately 10%. The positive colonies were then transferred to a second ‘indole master plate’ for future screening experiments against new substrates. In summary we believe this method is superior to alternative approaches based upon NAD(P)H monitoring assay in several respects, in particular, (i) since it is based upon screening colonies on agar plates it should be possible to evaluate a large number of mutants (ca. 105–106) and (ii) because the screen is for functional activity it eliminates the problem of identifying false positives due to uncoupling which is often observed.
Notes and references
- (a) M. Katagiri, B. N. Ganguli and I. C. Gunsalus, J. Biol. Chem., 1968, 243, 3543 CAS; (b) R. Raag, H. Li, B. C. Jones and T. L. Poulos, Biochem. J., 1993, 32, 4571 CrossRef CAS.
- O. Sibbesen, J. De Voss and P. R. Ortiz de Montellano, J. Biol. Chem., 1996, 271, 22462 CrossRef CAS.
- R. E. Speight, F. E. Hancock, C. Winkel, H. S. Bevinakatti, M. Sarkar, S. L. Flitsch and N. J. Turner, Tetrahedron: Asymmetry, 2004, 15, 2829 CrossRef CAS.
- K. E. O'Connor, A. W. Dobson and S. Hartmans, Appl. Environ. Microbiol., 1997, 63, 4287 CAS.
- R. W. Eaton and P. J. Chapman, J. Bacteriol., 1995, 177, 6983 CAS.
- (a) E. M. J. Gillam, L. M. Notley, D. Kim, R. G. Mundkowski, A. M. Aguinaldo, A. Volkov, F. H. Arnold, P. Soucek, J. DeVoss and F. P. Guengerich, Biochem. Biophys. Res. Commun., 1999, 265, 469 CrossRef CAS; (b) M. J. Elizabeth, L. M. Gillam, H. C. Notley, J. De Voss and F. P. Guengerich, Biochemistry, 2000, 39, 13817 CrossRef; (c) K. Nakamura, M. V. Martin and F. P. Guengerich, Arch. Biochem. Biophys., 2001, 395, 25 CrossRef CAS.
- L. Qing-Shan, U. Schwaneberg, P. Fischer and R. D. Schmid, Chem. Eur. J., 2000, 6, 1531 CrossRef CAS.
- A total of 24 P450CAM variants were selected for further analysis and sequencing from the Y96F98 library for the hydroxylation of diphenylmethane. A further 24 were selected with low activity (< 10% turnover). All of these variants were used to construct a ‘master plate’ of 48 P450CAM proteins which would be used for the primary screening experiment against new substrates (i.e. indole).
- (a) B. D. Ensley, B. J. Ratzkin, T. D. Osslund, M. J. Simon, L. P. Wackett and D. T. Gibson, Science, 1983, 222, 167 CrossRef CAS; (b) D. Murdock, B. D. Ensley, C. Serdar and M. Thalen, Bio/Technology, 1993, 11, 381 Search PubMed.
- A 400 ml LB medium with appropriate antibiotics inoculated with overnight culture of the mutants. The culture was grown to optical density of 0.4 (A600) at 30 °C. Induction was achieved using 0.02% of L-arabinose. After 30 minutes of induction, indole (1 mM final concentration) was delivered in DMSO (0.5%) into the medium. After 24 hours biotransformation at 30 °C, cells were harvested at 10 K rpm for 30 minutes. Cells then were washed with water. The residual blue pellet was extracted with tetrahydrofuran (THF, 3 × 25 ml). The solvent was evaporated to dryness and red pigment extracted from the residue with ethanol (5 × 10 ml). The ethanol extract was evaporated to dryness and a silica gel column used for separation and purification. (DC 60 Merck, eluted with THF ∶ petroleum ether 1 ∶ 2).
| This journal is © The Royal Society of Chemistry 2005 |
