Co-transport of H+/Cl− by a synthetic prodigiosin mimic†
Philip A.
Gale
*a,
Mark E.
Light
a,
Beth
McNally
b,
Korakot
Navakhun
a,
Kate E.
Sliwinski
a and
Bradley D.
Smith
*b
aSchool of Chemistry, University of Southampton, Southampton, UK SO17 1BJ. E-mail: philip.gale@soton.ac.uk; Fax: 44 2380596805; Tel: 44 23 80593332
bDepartment of Chemistry and Biochemistry, University of Notre Dame, IN 46556, USA. E-mail: smith.115@nd.edu; Fax: 1 574 631 6652; Tel: 1 574 631 8632
First published on 25th May 2005
Abstract
An amidopyrrole with appended imidazole group can bind and co-transport H+/Cl− across vesicle membranes much more effectively than an analogue with an appended pyridyl group.
A number of membrane-bound cellular compartments, such as the organelles of the biosynthetic and endocytic pathways, maintain acidic interiors that are crucial for organelle function and cell survival. There is literature evidence that connects abnormal changes in organelle steady-state pH with the pathology of several diseases.1 In addition, drug-resistant tumor cells are known to have organelles that are unusually acidic, and chemical agents that deacidify these organelles can restore drug-sensitivity.2,3 Deacidification of organelles can be induced by treatment with weak bases, ionophores, or v-type ATPase inhibtors. A subclass of the weak bases are compounds that act as H+/Cl− co-transporters. The best known examples are the prodigiosins, a family of naturally occurring pyrrole alkaloids,4 produced by microorganisms such as Streptomyces and Serratia.5,6 They have the general structure 1, and exhibit a range of potentially useful biological activities, including immunosuppression, induction of tumor cell apoptosis, and toxicity against bacteria, protozoa, fungi and malaria parasite.7,8 There is clear evidence that prodigiosins promote the co-transport of H+/Cl− across bilayer membranes.9–13 However, it is not conclusive which, if any, of the biological activities are directly due to organelle deacidification. For example, the prodigiosins are also known to affect mitogen-activated kinase signalling cascades,14 and to facilitate double-strand DNA cleavage in the presence of Cu(II) and O2.15
We have initiated a collaborative program to develop organic compounds that mimic the ability of the prodigiosins to promote H+/Cl− co-transport across bilayer membranes. A subsequent objective is to use these compounds to induce organelle deacidification, and see if this leads to biological activity. Since the mimics will not have DNA cleavage ability, they should be useful tools for elucidating the mode of biological action for the prodigiosins.
The development of synthetic chloride transporters is an emerging topic in supramolecular chemistry. Examples include cholapods (anion receptors containing a cholic acid backbone) that function as mobile carriers,16 and synthetic peptides that form transmembrane channels.17 In terms of H+/Cl− co-transport, the lone synthetic example is from the group of J. T. Davis and co-workers, who discovered that calix[4]arene and oligophenoxyacetamide derivatives are capable of ion channel conductance and co-transport of H+/Cl− through vesicle and cell plasma membranes.18
As a starting point, we decided to take design cues from the prodigiosin structure and produce relatively simple receptors containing two NH hydrogen bond donor groups (from an amidopyrrole19 group) and one basic group for protonation (an imidazole in the case of 2 and a pyridine group in the case of 3). As free bases, the receptors were expected to have weak chloride affinities, but they should form lipophilic HCl complexes in acid. Thus, the receptors were designed to co-transport H+/Cl− from an acidic phase through a bilayer membrane to a high pH interface where decomplexation occurs. To this end, compounds 2 and 3 were synthesised in 27 and 13% respective yields from the reaction of 5-methyl-1H-pyrrole-2-carboxylic acid ethyl ester20 with the aluminium amides21 formed from 1-methyl-2-aminomethylimidazole22 or 2-aminopyridine with trimethylaluminium in dichloromethane.
The anion binding affinities of receptors 2 and 3 were studied using 1H NMR titration techniques with tetrabutylammonium chloride in dichloromethane-d2 and the stability constants elucidated using the EQNMR computer program.23 The stability constants were found to be < 10 M−1 in both cases. Enhanced chloride binding by compound 2 upon protonation in acetonitrile-d3 solution was demonstrated by measuring the stability constant in the absence (60 M−1) and presence (397 M−1) of one equivalent of HPF6 at 298 K. Compound 3 showed only a weak interaction with chloride both in the absence and presence of HPF6 (< 10 M−1) under the same conditions.
X-ray crystal structures of 2 as the free base and as the HCl salt provided explanations for the difference in chloride affinities. X-ray quality crystals of 2 were obtained by slow evaporation of a dichloromethane/methanol solution of the receptor.‡ In the solid-state, the molecules form a series of hydrogen-bonded tapes. The repeat units are amidopyrrole dimers (connected by two pyrrole NH⋯O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C hydrogen bonds (N⋯O 2.8351(14)
Å)), which are associated via amide NH⋯N-imidazole hydrogen bonds (N⋯N 2.8837(15)
Å)
(Fig. 1).
C hydrogen bonds (N⋯O 2.8351(14)
Å)), which are associated via amide NH⋯N-imidazole hydrogen bonds (N⋯N 2.8837(15)
Å)
(Fig. 1).
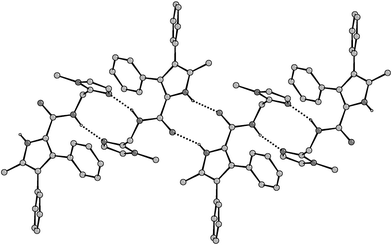 | ||
| Fig. 1 Crystal structure of compound 2 showing the formation of a hydrogen-bonded tape. | ||
Crystals of the HCl complex of compound 2 were obtained by slow evaporation of a dichloromethane/methanol/conc. HCl(aq) solution.§ The structure of 2·HCl revealed the formation of a ‘2 + 2’ dimer in the solid state with each chloride bound by three hydrogen bonds; two from the pyrrole (N⋯Cl 3.24(2) Å) and amide (N⋯Cl 3.29(2) Å) groups of one receptor and one from the imidazolium group of another (N⋯Cl 3.10(2) Å) (Fig. 2). With regard to the potential for membrane transport, a notable feature of the 2·HCl crystal structure is that all the polar and ionic functionality is inside the dimer; whereas, the exterior projects primarily lipophilic groups.
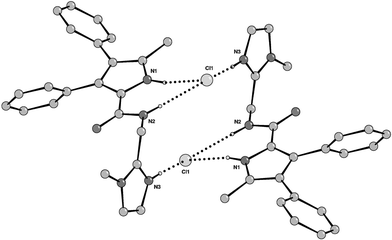 | ||
| Fig. 2 Crystal structure of HCl complex of 2 showing the formation of a ‘2 + 2’ hydrogen-bonded dimer. | ||
The abilities of receptors 2 and 3 to co-transport H+/Cl− across bilayer membranes were evaluated using unilamellar vesicles (200 nm mean diameter) composed of POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) ∶ cholesterol (7 ∶ 3 molar ratio). A chloride selective electrode16 was used to measure Cl− leakage from the vesicles under three different conditions, (a) inside vesicles: 500 mM NaCl, 5 mM citric acid, pH 4.0; outside vesicles: 500 mM NaNO3, 5 mM citric acid, pH 4.0, (b) inside vesicles: 500 mM NaCl, 5 mM citric acid, pH 4.0; outside vesicles: 500 mM NaNO3, 5 mM sodium phosphate, pH 6.7, (c) inside vesicles: 500 mM NaCl, 5 mM citric acid, pH 7.2; outside vesicles: 500 mM NaNO3, 5 mM citric acid, pH 7.2. As shown in Fig. 3, addition of receptor 2 (10 µM) induces Cl− release in a pH dependent manner. After a short induction period, a moderate rate of Cl− efflux was observed when the pH was 7.2 on both sides of the vesicle membrane, whereas no efflux was observed when both aqueous phases were acidic (pH 4.0). The highest efflux was observed when there was a pH gradient, that is, when the inside aqueous phase was acidic (pH 4.0) and the outside phase near neutral (pH 6.7). Furthermore, the initial flux with this pH gradient was found to increase in a linear fashion with the concentration of 2 (6 to 18 µM, data not shown). Receptor 3 was inactive as a chloride transporter under all of the above conditions (data not shown).
 | ||
| Fig. 3 Cl− efflux induced by the addition of 2 (10 µM) to vesicles containing NaCl (500 mM), X1 solution, and dispersed in NaNO3 (500 mM), X2 solution. ■ X1 and X2 = citric acid (5 mM), pH 4.0; ● X1 and X2 = citric acid (5 mM), pH 7.2; ▲ X1 = citric acid (5 mM), pH 4.0, X2 = sodium phosphate (5 mM), pH 6.7. At t = 1300 s, the vesicles were lysed to produce 100% Cl− release. | ||
Receptor 2 was subsequently tested for its ability to deacidify liposomes. The acid sensitive dye, Oregon Green® 514,24 was encapsulated inside POPC ∶ cholesterol (7 ∶ 3) vesicles at a pH of 4.0 (500 mM NaCl, 5 mM citric acid), and after dialysis to remove untrapped dye, the aqueous exterior phase was quickly adjusted to pH 7.2. Addition of 2 at t = 200 s induced an immediate discharge of the pH gradient, whereas, addition of compound 3 produced no enhancement of the background leakage rate (Fig. 4). The facilitated and background leakage of H+ shown in Fig. 4 appears to be substantially faster that the corresponding Cl− efflux shown in Fig. 3. However, this is an experimental artifact due to the very small fraction of aqueous phase that is entrapped inside the vesicles, and the large difference in probe sensitivities. The H+ detecting probe (Oregon Green® 514) is inside the vesicles and is quickly saturated by a very small amount of HCl efflux; whereas, the Cl− detecting probe (chloride selective electrode) is outside the vesicles and is much less sensitive to HCl appearance (it also has a slower reponse time).
 | ||
| Fig. 4 Change in Oregon Green® 514 fluorescence upon addition of: (A) 2 (10 µM), (B) 3 (8.8 µM), and (C) no receptor, at t = 200 s to vesicles containing Oregon Green (10 µM), NaCl (500 mM), and citric acid (5 mM), pH 4.0. The vesicles were dispersed in an external solution of NaNO3 (500 mM), and sodium phosphate (5 mM), pH 7.2. | ||
Taken together, the data are consistent with the transport model shown in Scheme 1. The free base of receptor 2 partitions into the vesicle membrane and diffuses to the interior membrane interface where it forms a lipophilic HCl complex (possibly the ‘2 + 2’ dimer in Fig. 2) that diffuses back through the membrane. Transport is not observed when the exterior phase is also acidic because 2·H+ is not sufficiently lipophilic to strongly partition from the bulk external aqueous into the vesicles. The pyridyl analogue 3 does not transport H+/Cl− out of vesicles, presumably because it is unable to form a kinetically active HCl complex.
 | ||
| Scheme 1 | ||
In summary, the amidopyrrole 2 with appended imidazole group can complex and co-transport H+/Cl− across vesicle membranes much more effectively than pyridyl analogue 3. The next stage of the research is to test if 2 and related analogues can induce organelle deacidification and see if this leads to prodigiosin-like biological activity.
PAG thanks Universities UK for an O.R.S. studentship to KN and the Royal Society for a University Research Fellowship. BDS acknowledges funding support from the NIH (USA). The authors thank the EPSRC for the crystallography and specifically W. Clegg for the synchrotron data collection, and Mike Hursthouse for access to the crystallographic facilities at the University of Southampton. We would like to thank Christopher J. Woods for the excellent cover illustration.
Notes and references
- O. A. Weisz, Traffic, 2003, 4, 57 CrossRef CAS.
- N. Altan, Y. Chen, M. Schindler and S. M. Simon, J. Exp. Med., 1998, 187, 1583 CrossRef CAS.
- Y. Chen, M. Schindler and S. M. Simon, J. Biol. Chem., 1999, 274, 18364 CrossRef CAS.
- A. Fürstner, Angew. Chem., Int. Ed., 2003, 42, 3582 CrossRef.
- N. N. Gerber, Crit. Rev. Microbiol., 1974, 3, 469 Search PubMed.
- J. W. Bennett and R. Bentley, Adv. Appl. Microbiol., 2000, 47, 1 Search PubMed.
- A. J. Castro, Nature, 1967, 213, 903 CAS.
- J. E. H. Lazaro, J. Nitcheu, R. Z. Predicala, G. C. Mangalindan, F. Nesslany, D. Marzin, G. P. Concepcion and B. Diquet, J. Nat. Toxins, 2002, 11, 367 Search PubMed.
- C. Yamamoto, H. Takemoto, H. Kuno, D. Yamamoto, A. Tsubura, K. Kamata, H. Hirata, A. Yamamoto, H. Kano, T. Seki and K. Inoue, Hepatology, 1999, 30, 894 CrossRef CAS.
- K. Tanigaki, T. Sato, Y. Tanaka, T. Ochi, A. Nishikawa, K. Nagai, H. Kawashima and S. Ohkuma, FEBS Lett., 2002, 524, 37 CrossRef CAS.
- T. Sato, H. Konno, Y. Tanaka, T. Kataoka, K. Nagai, H. H. Wasserman and S. Ohkuma, J. Biol. Chem., 1998, 273, 21455 CrossRef CAS.
- R. A. Gottlieb, J. Nordberg, E. Showronski and B. M. Babior, Proc. Natl. Acad. Sci. USA, 1996, 93, 654 CrossRef CAS.
- S. Ohkuma, T. Sato, M. Okamoto, H. Matsuya, K. Arai, T. Kataoka, K. Nagai and H. H. Wasserman, Biochem. J., 1998, 334, 731 CAS.
- R. Pérez-Tomás, B. Montaner and E. Llagostera, V. Soto-Cerrato Biochem. Pharmacol., 2003, 66, 1447 Search PubMed.
- R. A. Manderville, Curr. Med. Chem. Anti-Cancer Agents, 2001, 1, 195 Search PubMed.
- A. V. Koulov, T. N. Lambert, R. Shukla, M. Jain, J. M. Boon, B. D. Smith, H. Y. Li, D. N. Sheppard, J. B. Joos, J. P. Clare and A. P. Davis, Angew. Chem., Int. Ed., 2003, 42, 4931 CrossRef CAS.
- P. H. Schlesinger, R. Ferdani, J. Liu, J. Pajewska, R. Pajewski, M. Saito, H. Shabany and G. W. Gokel, J. Am. Chem. Soc., 2002, 124, 1848 CrossRef CAS.
- V. Sidorov, F. W. Kotch, J. L. Kuebler, Y.-F. Lam and J. T. Davis, J. Am. Chem. Soc., 2003, 125, 2840 CrossRef CAS; V. Sidorov, F. W. Kotch, G. Abdrakhmanova, R. Mizani, J. C. Fettinger and J. T. Davis, J. Am. Chem. Soc., 2002, 124, 2267 CrossRef CAS.
- P. A. Gale, Chem. Commun., 2005 10.1039/b504596g in this issue.
- R. J. Motekaitis, D. H. Heinert and A. E. Martell, J. Org. Chem., 1970, 35, 2504 CrossRef CAS.
- A. Basha, M. Lipton and S. M. Weinreb, Tetrahedron Lett., 1977, 18, 4171 CrossRef.
- K. J. Oberhansen, J. F. Richardson, R. M. Buchanan and W. Pierce, Polyhedron, 1989, 8, 659 CrossRef CAS; J. W. Canary, Y. Wang and R. Richard, Jr, Inorg. Synth., 1998, 32, 70 CAS.
- M. J. Hynes, J. Chem. Soc., Dalton Trans., 1993, 311 RSC.
- J. R. Lakowicz, H. Szmacinski and H. Lin, Anal. Biochem., 1999, 269, 162 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: experimental section. See http://www.rsc.org/suppdata/cc/b5/b503906a/index.sht |
| ‡ Crystal data for 2: Data collected on a Bruker Nonius KappaCCD equipped with a molybdenum rotating anode following standard procedures – C23H22N4O, Mr = 370.45, T = 120(2) K, triclinic, space group P-1, a = 7.3267(2), b = 10.4025(3), c = 13.6141(4) Å, α = 82.523(2), β = 76.069(2), γ = 82.898(2)°, V = 993.93(5) Å3, ρcalc = 1.238 g cm−3, μ = 0.078 mm−1, Z = 2, reflections collected: 17926, independent reflections: 3507 (Rint = 0.0702), final R indices [I > 2σI]: R1 = 0.0404, wR2 = 0.1053, R indices (all data): R1 = 0.0470, wR2 = 0.1099. CCDC 266941. See http://www.rsc.org/suppdata/cc/b5/b503906a/index.sht for crystallographic data in CIF or other electronic format. |
| § Crystal data for 2(HCl): Data collected on a Bruker SMART APEX2 CCD at station 9.8 of the Daresbury synchrotron – C23H25N4O2Cl, Mr = 424.92, T = 120(2) K, monoclinic, space group C2/c, a = 35.854(5), b = 6.249(5), c = 22.020(5) Å, β = 120.620(9)°, V = 4246(4) Å3, ρcalc = 1.33 g cm−3, μ = 0.208 mm−1, Z = 8, reflections collected: 12382, independent reflections: 3025 (Rint = 0.0689), final R indices [I > 2σI]: R1 = 0.0685, wR2 = 0.1693, R indices (all data): R1 = 0.0955, wR2 = 0.1872. CCDC 266942. See http://www.rsc.org/suppdata/cc/b5/b503906a/index.sht for crystallographic data in CIF or other electronic format. |
| This journal is © The Royal Society of Chemistry 2005 |


