Fluorescence studies of the intra-cellular distribution of zinc bis(thiosemicarbazone) complexes in human cancer cells†
Andrew R. Cowleya, Jason Davisa, Jonathan R. Dilworth*a, Paul S. Donnellya, Rachel Dobsona, Adrian Nightingalea, Josephine M. Peacha, Ben Shorea, David Kerrb and Len Seymourb
aChemistry Research Laboratory, University of Oxford, 12 Mansfield Road, Oxford, UK OX1 3TA. E-mail: jon.dilworth@chemistry.oxford.ac.uk
bDepartment of Clinical Pharmacology, University of Oxford, Radcliffe Infirmary, Woodstock Road, Oxford, UK OX2 6HE
First published on 21st January 2005
Abstract
The uptake of zinc bis(thiosemicarbazone) complexes in human cancer cells has been studied by fluorescence microscopy and the cellular distribution established, including the degree of uptake in the nucleus.
Thiosemicarbazones and their metal complexes have been known for many years to show a broad spectrum of therapeutic properties against a range of diseases, with antibacterial, antimalarial, antiviral and antitumour activities.1,2 Despite a sustained level of interest in the pharmacological properties of such complexes, details of the cellular distribution of these complexes are scarce, particularly in living cells. Certain zinc thiosemicarbazone complexes have been shown to be active as anti-tumour agents, are as cytotoxic as cisplatin and are also effective against cisplatin resistant cell lines.3 We have prepared a range of zinc bis(thiosemicarbazone) complexes and we here report the use of their intrinsic fluorescence to track their uptake in a range of human cancer cell types.
The zinc bis(thiosemicarbazone) complexes were prepared by a modification of the literature procedure4 by the reaction of zinc acetate with one equivalent of the ligand in ethanol under reflux. We have previously reported the dimeric structure of [Zn(ATSM)] (ATSM = diacetylbis(4-methyl-3-thiosemicarbazone)), in which each Zn(II) metal centre is in a square pyramidal environment with the apical site occupied by a sulfur from the neighbouring ligand.5 In DMSO the dimer becomes monomeric with an O-bound DMSO now in the fifth coordination site.‡ The low solubility of [Zn(ATSM)] in water prompted us to look at some different analogues.
Reaction of zinc chloride with diacetylbis(4-pyrrolidinyl-3-thiosemicarbazone) under the same conditions as above gives an unusual asymmetric complex: [ZnCl(ATSpyrH)].§ The zinc is again 5-coordinate, but now the apical ligand is chloride and one “limb” only of the bis(thiosemicarbazone) ligand is deprotonated, giving rise to an overall neutral complex. 1H NMR shows that the complex is fluxional at room temperature in d6-DMSO with the proton exchanging between the two thiosemicarbazone “limbs”. In aqueous solution, the apical chloride is replaced by a water molecule to give a much more soluble cationic species. An ORTEP representation of the structure¶ appears in Fig. 1, together with selected bond lengths. These confirm the asymmetric nature of the two “limbs” of the ligand.
![ORTEP representation of the structure of [ZnCl(ATSpyrH)]. Selected bond lengths (Å): Zn(1)–Cl(1), 2.2910(9); Zn(1)–S(1), 2.4092(9); Zn(1)–S(2), 2.3548(9); Zn(1)–N(2), 2.194(3); Zn(1)–N(3), 2.107(3); S(1)–C(1), 1.708(3); S(2)–C(4), 1.748(4); C(1)–N(1), 1.369(4); C(4)–N(4), 1.335(4); N(1)–H(1), 0.76(4).](/image/article/2005/CC/b417206j/b417206j-f1.gif) | ||
| Fig. 1 ORTEP representation of the structure of [ZnCl(ATSpyrH)]. Selected bond lengths (Å): Zn(1)–Cl(1), 2.2910(9); Zn(1)–S(1), 2.4092(9); Zn(1)–S(2), 2.3548(9); Zn(1)–N(2), 2.194(3); Zn(1)–N(3), 2.107(3); S(1)–C(1), 1.708(3); S(2)–C(4), 1.748(4); C(1)–N(1), 1.369(4); C(4)–N(4), 1.335(4); N(1)–H(1), 0.76(4). | ||
The fluorescence of zinc thiosemicarbazone complexes has only very recently been reported, and the photophysics of complexes of the type [Zn(4-Me2NC6H4CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NNCSNH2)2] have been interpreted in terms of intraligand excitation.6 The bis(thiosemicarbazone) complexes used here show emissions in the range 500–600 nm (see Fig. 2) dependent on ligand and solvent. The quantum yields for 0.01 mM DMSO solutions of [Zn(ATSM)] and [ZnCl(ATSpyrH)] are 1.25 × 10−3 and 2.31 × 10−3 respectively (each value ± 10%). Importantly, the fluorescence profile allows for a relatively low energy excitation that reduces potential interference from other fluorescent molecules in the cell.
NNCSNH2)2] have been interpreted in terms of intraligand excitation.6 The bis(thiosemicarbazone) complexes used here show emissions in the range 500–600 nm (see Fig. 2) dependent on ligand and solvent. The quantum yields for 0.01 mM DMSO solutions of [Zn(ATSM)] and [ZnCl(ATSpyrH)] are 1.25 × 10−3 and 2.31 × 10−3 respectively (each value ± 10%). Importantly, the fluorescence profile allows for a relatively low energy excitation that reduces potential interference from other fluorescent molecules in the cell.
![Emission spectra for 0.01 mM solutions of [Zn(ATSM)] and [ZnCl(ATSpyrH)] in DMSO. λex
= 410 nm.](/image/article/2005/CC/b417206j/b417206j-f2.gif) | ||
| Fig. 2 Emission spectra for 0.01 mM solutions of [Zn(ATSM)] and [ZnCl(ATSpyrH)] in DMSO. λex = 410 nm. | ||
For the measurements of cell uptake and speciation, samples of cells were grown in Petri dishes equipped with an aperture covered with a circular microscope slide coated with polylysine to ensure cell adhesion. A solution of the zinc complex (0.8 mL of 0.01 mM solution in 5% DMSO–water) was added to the cell suspension and the images shown below were all taken one hour after addition. A brightfield image of each cell was taken immediately prior to the irradiation and fluorescence measurement (Figs. 3–6). The cells photographed were chosen for picture quality, but are representative of the response observed across the entire treated population. The morphologies of the cells were entirely consistent with their having maintained viability. The black and white fluorescence images were then exported into ImageJ7 and scaled in colour. Controls involving the addition of a zinc salt or bis(thiosemicarbazone) ligand alone showed no fluorescence above a very low intensity background. This demonstrates that the observed fluorescence must be due to the presence of the intact zinc bis(thiosemicarbazone) complex.
![Left: an IGROV ovarian cancer cell. Right: a fluorescence image of the same cell after exposure to [Zn(ATSM)].](/image/article/2005/CC/b417206j/b417206j-f3.gif) | ||
| Fig. 3 Left: an IGROV ovarian cancer cell. Right: a fluorescence image of the same cell after exposure to [Zn(ATSM)]. | ||
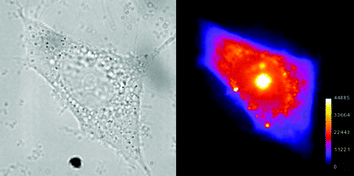 | ||
| Fig. 4 As Fig. 3 but with MCF-7 breast cancer cells. | ||
![PC-3 prostate cancer cells with [Zn(ATSM)].](/image/article/2005/CC/b417206j/b417206j-f5.gif) | ||
| Fig. 5 PC-3 prostate cancer cells with [Zn(ATSM)]. | ||
![PC-3 prostate cancer cells with [ZnCl(ATSpyrH)].](/image/article/2005/CC/b417206j/b417206j-f6.gif) | ||
| Fig. 6 PC-3 prostate cancer cells with [ZnCl(ATSpyrH)]. | ||
In each case there is evidence for significant zinc complex uptake within the cells with, as might be expected, marked differences depending on the cell type and the nature of the zinc complex. For IGROV ovarian cancer cells and [Zn(ATSM)] (Fig. 3), there is a low general uptake in the cytosol but distinct higher concentrations within organelles in the cells. We believe from acridine orange staining experiments that these are lysosomes and the low pH generally found in these would be consistent with a protonation enhanced trapping mechanism. Protonation of the complexes results in positive charge which would prevent cell or organelle washout. The images also clearly show uptake in the nucleolus. Independent fluorescence and UV–visible measurements of the binding of [Zn(ATSM)] to calf thymus DNA and competition experiments with ethidium bromide suggest strongly an electrostatic rather than intercalative interaction and may be relevant to the observed nucleolar uptake. The intermolecular hydrogen bonding observed in the solid state structures indicates that such interactions are feasible (N(6)⋯N(4)′ = 3.048(3) Å) (see ESI†)
Fig. 4 shows the same experiment with MCF-7 breast cancer cells. Here the cytoplasmic distribution is more uniform and take up in the nucleolus is significantly greater.
Finally in the case of PC-3 prostate cancer cells (Figs. 5 and 6), the distribution is again different, with the zinc complexes appearing to be concentrated in a relatively small zone of the cell with a low general uptake in the rest of the cytoplasm. Significantly the pyrrolidine substituted complex shows virtually no uptake in the nucleolus as compared to [Zn(ATSM)].
These results present the first fluorescence measurements of the uptake and distribution of thiosemicarbazone complexes in living cells. They show clearly that uptake in the nucleus is a sensitive function of the terminal nitrogen substituents on the complexes and that the integrity of the complexes is maintained. It also demonstrates that the distribution within the cytoplasm depends on the cell type. We have reported the dependence of cell uptake of metal complexes on cell type8 and this work provides a further example. Fluorescence microscopy offers a method of establishing Structure–Activity Relationships for zinc complexes in cells and thereby optimising the therapeutic activity. We also note that the structures of the zinc complexes are very similar to their copper analogues and that the cell uptake mechanisms may well be similar. We are currently investigating the uptake of zinc complexes of tridentate thiosemicarbazones and also rhenium complexes containing the tricarbonyl core which also have intrinsic fluorescence, and probing the mechanisms of uptake.
We are indebted to Dr S. Faulkner of the University of Manchester for helpful discussions on the photophysics of the zinc complexes and Mrs S. Phipps from the Department of Clinical Pharmacology, University of Oxford, for providing us with cells.
Notes and references
- D. X. West, S. B. Padhye and P. B. Sonawane, Struct. Bonding, 1991, 76, 1 CAS.
- D. X. West, A. E. Liberta, S. B. Padhye, R. C. Chikate, P. B. Sonawane, A. S. Kumbhar and R. G. Yerande, Coord. Chem. Rev., 1993, 123, 49 CrossRef CAS.
- H. Beraldo and D. Gambino, Mini-Rev. Med. Chem., 2004, 4, 31 Search PubMed.
- N. C. Kasuga, K. Sekino, M. Ishikawa, A. Honda, M. Yokoyama, S. Nakano, N. Simada, C. Koumo and K. Nomiya, J. Inorg. Biochem., 2003, 96, 298 CrossRef CAS.
- A. R. Cowley, J. R. Dilworth, P. S. Donnelly, E. Labisbal and A. J. Sousa, J. Am. Chem. Soc., 2002, 124, 5270 CrossRef CAS.
- Z.-M. Xue, Y.-P. Tian, D. Wang and M. H. Jiang, Dalton. Trans., 2003, 1373 RSC.
- W. S. Rasband, ImageJ, National Institutes of Health, Bethesda, Maryland, USA, http://rsb.info.nih.gov/ij/, 1997–2004.
- M. A. Stalteri, S. J. Parrot, V. A. Griffiths, J. R. Dilworth and S. J. Mather, Nucl. Med. Commun., 1997, 18, 870 CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: further details of the crystallographic study. See http://www.rsc.org/suppdata/cc/b4/b417206j/ |
| ‡ Crystal data: C12H26N6O2S4Zn, M = 480.00, monoclinic, a = 17.1632(5), b = 8.1574(3), c = 17.4311(6) Å, β = 120.6784(13), U = 2098.9 Å3, T = 150 K, space group P 21/c, Z = 4, μ = 1.586 mm−1, 19382 reflections measured, 5095 unique (Rint = 0.057) which were used in all calculations. R = 0.0348, wR [I > 3σ(I)] = 0.0407. CCDC 256169. See http://www.rsc.org/suppdata/cc/b4/b417206j/ for crystallographic data in .cif format. |
| § A solution of ZnCl2 (2.94 mmol, 0.401 g) in 30 mL ethanol was added dropwise to a solution of diacetylbis(4-pyrrolidinyl-3-thiosemicarbazone) (2.94 mmol, 1 g) in 30 mL chloroform. The reaction was refluxed for 4 hours and then cooled to room temperature. The yellow precipitate was collected by filtration, triturated with hot ethanol, filtered and washed with diethyl ether. This was then dried under vacuum to give a bright yellow powder. (2.27 mmol, 0.994 g, 77% yield) 1H NMR (d6-DMSO, 300 MHz) δ 1.90 (s, 8H, NCH2CH2), 2.30 (s, 6H, CH3), 3.70 (s, 8H, NCH2). ES MS (negative ion): m/z 437 = [ZnCl(ATSpyr)]−, (positive ion): m/z 403 = [Zn(ATSpyrH)]+, m/z 809 = {[Zn(ATSpyrH2)][Zn(ATSpyrH)]}+, m/z 1213 = {2[Zn(ATSpyrH2)] [Zn(ATSpyrH)]}+. FAB (positive ion): m/z 403 = [Zn(ATSpyrH)]+. UV–visible and fluorescence spectra were measured with a Perkin Elmer Lamda 19 spectrometer and a Hitachi F-4500 fluorescence spectrophotometer respectively. HPLC-grade solvents were used at all times. Cellular images were taken using a Nikon TE-2000E microscope with an Andor iXon iCCD camera. Irradiation was carried out with filtered radiation from a mercury lamp at ca. 480 nm and the excitation time was kept as short as possible to minimize photobleaching. The measurements were carried out for at least ten cells in each Petri dish then repeated for different preparations of each cell type and were found to be reproducible. |
¶ Crystal data: C14H23ClN6S2Zn, M
= 440.33, triclinic, a
= 7.8934(3), b
= 8.5558(3), c
= 14.1976(5)
Å, α
= 74.5790(15), β
= 87.2903(16), γ
= 85.2071(19)
°, U
= 920.76(6)
Å3, T
= 150 K, space group P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) , Z
= 2, μ
= 1.715 mm−1, 16265 reflections measured, 4209 unique (Rint
= 0.074) which were used in all calculations. R
= 0.0347, wR
[I > 3σ(I)]
= 0.0396. CCDC 256170. See http://www.rsc.org/suppdata/cc/b4/b417206j/ for crystallographic data in .cif format. , Z
= 2, μ
= 1.715 mm−1, 16265 reflections measured, 4209 unique (Rint
= 0.074) which were used in all calculations. R
= 0.0347, wR
[I > 3σ(I)]
= 0.0396. CCDC 256170. See http://www.rsc.org/suppdata/cc/b4/b417206j/ for crystallographic data in .cif format. |
| This journal is © The Royal Society of Chemistry 2005 |
