Semiconductor quantum dots and free radical induced DNA nicking†
Mark
Green
*a and
Emily
Howman
b
aDepartment of Physics, King's College London, Strand, London, UK WC2R 2LS. E-mail: mark.a.green@kcl.ac.uk; Fax: 020 7848 2420; Tel: 020 7848 2121
bDepartment of Human Anatomy and Genetics, University of Oxford, South Parks Road, Oxford, UK OX1 3QX
First published on 25th November 2004
Abstract
There is a growing interest in the use of semiconductor quantum dots as fluorescent markers in biological applications.1 However, there are concerns regarding the potential environmental impact and toxic nature of these nanomaterials.2 In this study, we have investigated the interaction of water-soluble semiconductor quantum dots with supercoiled DNA.
To date, investigations into the adverse biological effects of nanomaterials have centered on fullerenes3 and titania.4 A recent study by Derfus et al. highlighted the cytotoxicity of CdSe and CdSe/ZnS quantum dots and attributed the detrimental effects to free cadmium ions in solution.5 If semiconductor quantum dots are to be used routinely in in vivo studies or in DNA based assays, it is essential to know that the materials themselves do not induce adverse effects or cause misleading results. In this paper, we report the observation of DNA damage in plasmid nicking assays with water-soluble CdSe/ZnS quantum dots. We also suggest a nicking mechanism based on observed free radical generation.
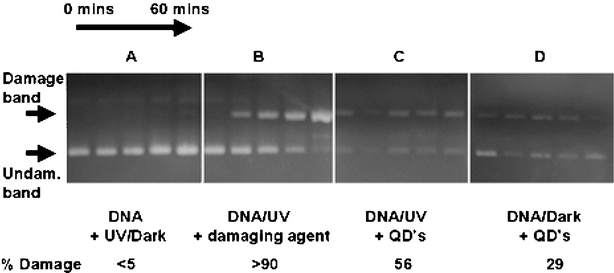 | ||
| Fig. 1 Plasmid nicking assays of supercoiled DNA. From left to right shows aliquots taken after every 15 min (0–60 min). (A) DNA after UV excitation/dark. All show supercoiled DNA undamaged; (B) DNA incubated with a known plasmid nicking agent. Shows change from undamaged (0 min) to almost complete damage (60 min); (C) DNA after incubation with quantum dots and UV excitation, shows an almost constant level of damage; (D) DNA after incubation with quantum dots in the dark, shows a slightly weaker damaged band. The figure is reproduced in more detail in ESI†. | ||
Commercially available water-soluble quantum dots (cadmium selenide capped with a shell of zinc sulfide, complete with biotin surface functionality) were incubated with supercoiled double strands of DNA (which are highly sensitive to free radicals) in the dark and under UV excitation (see ESI†). A single free radical can break a deoxyribose unit that leads to the uncoiling of the DNA strand.6
Following a set period of incubation (between 0 and 60 min, 15 min intervals), the DNA was precipitated from the quantum dots solution and then run on an electrophoresis gel, where supercoiled and uncoiled (damaged) DNA can be detected at different positions. Failure to isolate the DNA from the dots resulted in unresolved bands.
Blank assays with DNA (UV and dark) showed undamaged bands of DNA (Fig. 1a). Samples of DNA incubated with a known plasmid nicking agent (titania, P25) were also run to ascertain the position of the damaged band of DNA (Fig. 1b).4 Other, faint bands can be observed in the samples showing DNA damage; this is consistent with DNA strands cleaved at multiple sites. Finally, assays with DNA that had been incubated with quantum dots prior to isolation were run to determine if the quantum dots damaged the DNA.
Assays with DNA that had been incubated with quantum dots and exposed to UV light showed damage (Fig. 1c). Samples of DNA incubated with quantum dots in the dark also showed evidence of a damaged band (Fig. 1d) not observed in assays using DNA stored alone in the dark. It is worth noting that the intensity of the bands in assays carried out with DNA that were incubated with quantum dots are significantly weaker than experiments run without quantum dots present. We attribute this to the DNA coordinating to the dots during incubation, resulting in smaller yields of DNA when isolated. We estimate up to 70% of the DNA coordinated to the dots non-specifically and was therefore unavailable for assay analysis.
It is important to note that plasmid damage was observed in assays run with DNA isolated from dots at time 0, indicating immediate modification upon mixing the dots and the DNA. Tricosanthin coupled dots have previously been shown to nick plasmid DNA; however, tricosanthin is also known to nick DNA and was assumed to be the origin of the cleavage.7
The intensities of the damaged bands were calculated; DNA incubated with dots and exposed to UV light displayed ca. 56% modified (damaged) DNA. Assays run with DNA that had been incubated with quantum dots in the dark revealed less DNA damage (29%) suggesting that the major damaging factor is not necessarily UV induced, unlike DNA damage observed with other semiconductor particles such as TiO2.4 This strongly suggests that the damaging mechanism for DNA incubated with quantum dots is not a simple photo-induced free radical process.
DNA nicking from aqueous dispersions of the TiO2 has previously been attributed to the generation of singlet oxygen and hydroxyl free radicals by photogenerated charge carriers.4 In these cases, photoactivation (generation of electrons and holes) was required. Photoactivated semiconductor quantum dots are also expected to generate similar free radicals when in aqueous solution; hence a similar mechanism for DNA damage would be expected. The ZnS shell only confines the hole to the core of the quantum dot, whilst the electron extends over the entire structure. Thus, whilst protecting the emitting core from oxidation, the ZnS shell should not inhibit electron induced free radical generation in water.8
In the present study comprising water-soluble semiconductor quantum dots, another mechanism must also be invoked as DNA damage is also evidenced in the dark. In the case of semiconductor quantum dots, we suggest a sulfur related free radical species may also contribute.9,10 Previous work by Bowen-Katari et al. has shown that CdSe quantum dots slowly photo-oxidise over 24 h to yield selenium dioxide that slowly dissociate from the particle over a 96 h period.11
In the case of water soluble core/shell semiconductor quantum dots, we suggest the ZnS cap slowly oxidises in the presence of air and water (the aqueous samples are supplied and stored under ambient conditions) giving SO2 that desorbs into solution, producing the free radical SO2−˙.12,13 The oxidation of ZnS is a reaction commonly used in industrial processes.14 Likewise, the photodegradation of CdS nanoparticles and associated release of free radicals in aqueous solution is well documented.15,16 The SO2−˙ free radical has been shown previously to air-oxidise to form superoxide,12 which can produce hydroxyl free radicals that are known to nick plasmid DNA.17,18 The direct modification of nucleic acids by sulfoxide radicals has also been reported.9,10
The slow oxidation and gradual desorption of surface oxide into solution can therefore explain why DNA damage is observed when the sample is stored in the dark having previously been stored in ambient conditions. To test this hypothesis, free radical spin trap experiments were carried out on samples prepared without any bioactive species on the surface or unknown agents in the supplied buffer solution, to eliminate potentially unknown sources of free radicals. CdSe quantum dots capped with a ZnS shell were prepared by organometallic methods under an inert atmosphere and the entire structure passivated with tri-n-octylphosphine oxide and tri-n-octylphosphine. The surface ligand was then exchanged with 11-mercaptoundecanoic acid under an inert atmosphere and the carboxylic acid group deprotonated using potassium butoxide in DMF, giving water soluble, luminescent CdSe/ZnS quantum dots (see ESI†).
The sample was then dissolved in deionised water with spin trap DMPO (5,5-dimethyl-1-pyrroline N-oxide) and left for 72 h in ambient conditions. Samples were then excited with a solar simulator (1 min), and an electron spin resonance (ESR) signal consistent with the presence of free radicals was observed (Fig. 2). Control ESR experiments were also conducted on samples in the dark (i.e., in the absence of excitation) which importantly showed signals confirming free radical generation, although the signal was weaker (Fig. 2). Despite confirming the presence of free radicals in both illuminated samples and samples retained in the dark, we cannot distinguish between the two free radical species easily and experiments are underway to elucidate the identity of the generated species and to determine the ratio between the pair.
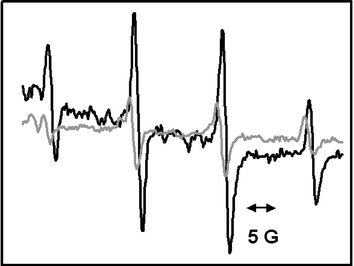 | ||
| Fig. 2 ESR spectrum of water-soluble CdSe/ZnS quantum dots and a spin trap (DMPO) confirming the presence of free radicals in solution. The dark line spectrum represents an illuminated sample; the grey line spectrum represents a sample in the dark. | ||
There are, however, potential methods of avoiding such free radicals. Doped materials (for example, Mn2+ doped into a lattice) could hold charge carriers in the internal structure of the dots. Development of new core/shell structures may completely confine charge carriers to the core of the material. III–V quantum dots have stable surface oxides that are unlikely to leach into solution. Incorporation of quantum dots into beads may isolate the material from the biological environment. It is also worth noting that although the quantum dots do damage DNA, the effect is not as severe as damage caused by titania (P25, control) under similar conditions.
In conclusion, we have demonstrated that water-soluble II–VI core/shell semiconductor quantum dots can nick DNA and we attribute the observation to free radicals, both photogenerated and surface oxide generated. These results suggest that there may be serious issues to address concerning the use of such materials in DNA based assays or in in vivo applications, as well as highlighting potential toxicological and environmental implications.
Note and references
- A. P. Alivisatos, Nature Biotechnol., 2004, 22, 47 CrossRef.
- V. L. Colvin, Nature Biotechnol., 2003, 21, 1166 CrossRef CAS.
- C. M. Sayes, J. D. Fortner, W. Guo, D. Lyon, A. M. Boyd, K. D. Ausman, Y. J. Tao, B. Sitharaman, L. J. Wilson, J. B. Hughes, J. L. West and V. L. Colvin, Nano Lett., 2004, 4, 1881 CrossRef CAS.
- G. Wakefield, S. Lipscomb, E. Holland and J. Knowland, Photochem. Photobiol. Sci., 2004, 3, 648 RSC.
- A. M. Derfus, W. C. W. Chan and S. N. Bhatia, Nano Lett., 2004, 4, 11 CrossRef CAS.
- E. Damiani, L. Greci, R. Parsons and J. Knowland, Free Radical Biol. Med., 1999, 26, 809 CrossRef CAS.
- Z. Chunyang, M. Hui, S. Nie, D. Yao, J. Lei and C. Dieyan, Analyst, 2000, 125, 1029 RSC.
- B. O. Dabbousi, J. Rodriguez-Viejo, F. V. Mikulec, J. R. Heine, H. Mattoussi, R. Ober, K. F. Jensen and M. G. Bawendi, J. Phys. Chem. B., 1997, 101, 9463 CrossRef CAS.
- H. Hayatsu, Prog. Nucleic Acid Res. Mol. Biol., 1976, 16, 75 Search PubMed.
- H. Hayatsu and R. C. Miller, Biochem. Biophys. Res. Commun., 1972, 46, 120 CrossRef CAS.
- J. E. Bowen-Katari, V. L. Colvin and A. P. Alivisatos, J. Phys. Chem., 1994, 98, 4l09.
- C. Mottley, L. S. Harman and R. P. Mason, Biochem. Pharmacol., 1985, 34, 3005 CrossRef CAS.
- S. Gümüşlü, D. Kipmen, S. Bilmen, P. Yargicoğlu and A. Ağar, Ind. Health, 2002, 38, 319.
- F. Patisson and D. Ablitzer, Powder Technol., 2002, 128, 3000 CrossRef.
- A. Henglein, Ber. Bunsen-Ges. Phys. Chem., 1982, 86, 301 CAS.
- J. R. Harbour and M. L. Hair, J. Phys. Chem., 1977, 81, 1791 CrossRef CAS.
- Y. Yamamoto, J. Dermatol. Sci., 2001, 27, S1 CrossRef CAS.
- G. Wakefield, M. Green, S. Lipscomb and B. Flutter, Mater. Sci. Technol., 2004, 20, 985 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Details of Fig. 1, plasmid nicking assays, spin-trap experiments, nanoparticle synthesis and phase transfer. See http://www.rsc.org/suppdata/cc/b4/b413175d/ |
| This journal is © The Royal Society of Chemistry 2005 |
