Controlled formation of biosilica structures in vitro†
Rajesh R.
Naik
a,
Patrick W.
Whitlock
ab,
Francisco
Rodriguez
a,
Lawrence L.
Brott
a,
Diana D.
Glawe
ac,
Stephen J.
Clarson
b and
Morley O.
Stone
*a
aMaterials and Manufacturing Directorate, Air Force Research Laboratory, Wright-Patterson Air Force Base, Dayton, OH 45433, USA
bDepartment of Materials Science and Engineering, University of Cincinnati, Cincinnati, OH 45221, USA
cDepartment of Engineering Science, Trinity University, San Antonio, TX 78212, USA. E-mail: Morley.Stone@wpafb.af.mil
First published on 11th December 2002
Abstract
Herein we describe the controlled formation of biosilica structures by manipulation of the physical reaction environment; we were able to synthesize arched and elongated silica structures using a synthetic peptide; the results presented here are evidence that in vitro biocatalysis may be controlled in order to form desired silica structures.
Numerous examples of nanopatterning and nanostructure are commonly found in nature, most apparent in the marine diatoms and sponges.1,2 These organisms are able to form intricate siliceous nanostructures within their cell walls with precise control. The structures are reproduced generation after generation with great fidelity.2 Proteins known as silicateins isolated from within sponge silica have been shown to catalyze the in vitro polymerization of silica from tetraethoxysilane at neutral pH.3,4 Silaffins, a set of cationic polypeptides isolated from the diatom Cylindrotheca fusiformis, can generate a network of silica nanospheres when added to a solution of silicic acid in vitro.5 The diatom cell walls are considered as a paradigm for the controlled production of nanostructure silica. The conventional chemical synthesis of silica-based materials requires harsh conditions such as extreme temperature, pH and pressure, whereas biosilification occurs at ambient temperature and pressure. The low cost, low impact, biomimetic formation of silica nanostructures using polypeptides overcomes all of these limitations. More importantly, biosilification allows control of morphology at the structural level desired for advanced nano-applications. Sil1 protein from C. fusiformis is composed of seven highly homologous repeating units (R1 to R7). We, as well as others, have observed the formation of silica nanospheres using a 19 amino-acid R5 unit peptide [H2N-SSKKSGSYSGSKGSKRRIL-COOH] of the Sil1 protein.5,6
Here we report the formation of silica morphologies that are driven by the flow dynamics of the enzymatic reaction. In a typical procedure, 1 M tetramethoxysilane (TMOS) dissolved in 1 mM hydrochloric acid was mixed with the Sil1-derived R5 peptide at a concentration of 10 mg ml−1 and the reaction was incubated for 2–5 min.6 The silica precipitate obtained was analyzed by scanning electron microscopy (SEM). Silica spheres obtained from the TMOS reaction are shown in Fig. 1A. In the absence of R5, no precipitate was observed and the reaction mixture formed a gel within 24 h. Silica pecipitate was not obtained in the absence of R5 peptide or when a non-specific peptide was used in the above reaction. When a stream of nitrogen was slowly passed through the reaction vessel, an arch-like structure was formed. As the nitrogen gas was passed through the reaction mixture, contact between newly formed bubbles resulted in the formation of an interface. We propose that the arched morphology is a result of the continual addition of silica spheres to the growing structure, (Fig. 1B and C). Incorporated silica, similar in diameter (∼500 nm) to that produced under ambient conditions with the R5 peptide and TMOS, can be easily identified along the length of these structures. It is important to appreciate that even though aggregation of silica spheres is a regularly observed aspect of diatom silica polymerization,7,8 directional growth to form elongated silica strands is required to form larger three-dimensional structures.9,10 Directional growth occurs in a microtubule-dependent manner, which has been proposed to localize actively polymerizing silica centers at the edge of the silica deposition vesicle (SDV) where polymerization occurs.11,12 We propose that a similar, albeit less complex, mechanical situation may have been created in vitro, resulting in the observed morphology.
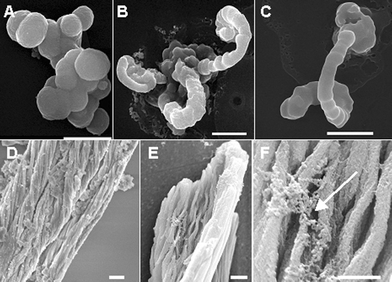 | ||
| Fig. 1 Scanning electron micrographs of biosilica structures obtained using the R5 peptide. (A) Spherical silica structures obtained under static reaction conditions. (B) and (C): Arch-shaped morphologies were obtained by bubbling nitrogen gas through the reaction mixture. (D) and (E): Fibrillar morphologies were obtained by applying a mechanical shear to the reaction mixture. (F) Rough agglomerates of silica spheres (arrow). Scale bar 1 μm. | ||
Next, we applied shear to a constant, linear, microvolume of the R5/TMOS reaction mixture by cycling the reaction mixture back and forth inside a Tygon® tube (1.6 mm inner diameter), similar to a ‘slug flow’. Elongated fibers were formed as a result of the applied shear (Fig. 1D and E). Several levels of linear organization characterize this structure. Rough agglomerates of silica spheres appear to constitute the primary structural organization (Fig. 1F). The microfibrils (100–300 nm in diameter) make up the larger, fibrillar morphology observed at lower magnification. Electron dispersive spectroscopy (EDS) analysis of all of the above morphologies produced high silicon and oxygen content and limited carbon content as would be expected from silica. No microfibrils were observed when a control reaction (lacking R5 peptide or using a non-specific peptide) was subjected to shear as described above. In addition, we and others have observed that the R5 as well as other silica-condensing peptides are tightly associated with the silica structure (see ESI†).5
We have shown several silica nano-morphologies catalyzed by the R5 peptide of C. fusiformis under varied reaction conditions. These structures range from common spheres to highly organized and complex fibrillar morphologies, which display remarkable organization at the nanometer size-scale. Through careful manipulation of the environment and the use of mechanical force, we were able to direct the formation of silica in such a way as to produce a desired morphology. The results presented here show that by simply moving the silification site in a directed manner during the course of polymerization results in an altered morophology. Silica spheres are the lowest free energy structures formed in a static environment, but elongated strands occur when the polymerization front is in motion. In nature, marine diatoms and sponges are able to direct synthesis of complex structures within their cell walls via complex molecular methods that control the transport and deposition of silica.3–5,12,13 The means employed herein also appear to direct the transport and deposition of silica as it is formed from the silane precursor by the R5 peptide. Application of a linear shear force and the presence of an interfacial reaction surface both affect the deposition of silica, producing separate and distinct morphologies that are unlike the spheres observed under normal reaction conditions. The localized transport phenomenon occurring at the interface between the tube walls and the surrounding fluid may be partly responsible for the formation of these elongated fibrillar morphologies. We are currently investigating the mechanism involved in generating these novel morphologies and developing new methods to control the deposition of silica for various applications.
This work was funded by the Air Force Office of Scientific Research.
Notes and references
-
F. E. Round, R. M. Crawford and D. G. Mann, The Diatoms: Biology & Morphology of the Genera,Cambridge University Press, Cambridge, 1990 Search PubMed
.
-
S. Mann, Biomineralization: Principles and Concepts in Bioinorganic Materials Chemistry, Oxford University Press, 2001 Search PubMed
.
- K. Shimizu, J. Cha, G. D. Stucky and D. E. Morse, Proc. Natl. Acad. Sci. USA, 1998, 95, 6234 CrossRef CAS
.
- J. N. Cha, K. Shimizu, Y. Zhou, S. C. Christiansen, B. F. Chmelka, G. D. Stucky and D. E. Morse, Proc. Natl. Acad. Sci. USA, 1999, 96, 361 CrossRef CAS
.
- N. Kroger, R. Deutzmann and M. Sumper, Science, 1999, 286, 1129 CrossRef CAS
.
- L. L. Brott, R. R. Naik, D. J. Pikas, S. M. Kirkpatrick, D. W. Tomlin, P. W. Whitlock, S. J. Clarson and M. O. Stone, Nature, 2001, 413, 291 CrossRef CAS
.
- M. L. Chiappino and B. E. Volcani, Protoplasma, 1977, 93, 205 Search PubMed
.
- A-M. M. Schmid and D. Schulz, Protoplasma, 1979, 100, 267 Search PubMed
.
- A. M. M. Schmid and B. E. Volcani, J. Phycol., 1983, 19, 387 CrossRef
.
-
J. Pickett-Heaps, A. M. M. Schmid and L. A. Edgar, in Progress in Phycological Research, ed. F. E. Round and D. J. Chapman, Biopress, Bristol, UK, 1990, vol. 7, p. 1 Search PubMed
.
- R. W. Drum and H. S. Pankratz, J. Ultrastruct. Res., 1964, 10, 217 Search PubMed
.
- S. A. Crawford, M. J. Higgins, P. Mulvaney and R. Wetherbee, J. Phycol., 2001, 37, 1 CrossRef
.
- M. Hildebrand, K. Dahlin and B. E. Volcani, Mol. Gen. Genet., 1998, 260, 480 Search PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available: incorporation of silica-condensing peptide into the silica structure. See http://www.rsc.org/suppdata/cc/b2/b210635c/ |
| This journal is © The Royal Society of Chemistry 2003 |
