Simple phenylpropanoids: recent advances in biological activities, biosynthetic pathways, and microbial production†
Zhanpin
Zhu
 a,
Ruibing
Chen
a,
Ruibing
Chen
 *a and
Lei
Zhang
*a and
Lei
Zhang
 *abc
*abc
aDepartment of Pharmaceutical Botany, School of Pharmacy, Naval Medical University, Shanghai 200433, China. E-mail: rbchenstar@163.com; nmu_dpb@aliyun.com
bInstitute of Interdisciplinary Integrative Medicine Research, Medical School of Nantong University, Nantong 226001, China
cInnovative Drug R&D Centre, College of Life Sciences, Huaibei Normal University, Huaibei 235000, China
First published on 9th October 2023
Abstract
Covering: 2000 to 2023
Simple phenylpropanoids are a large group of natural products with primary C6–C3 skeletons. They are not only important biomolecules for plant growth but also crucial chemicals for high-value industries, including fragrances, nutraceuticals, biomaterials, and pharmaceuticals. However, with the growing global demand for simple phenylpropanoids, direct plant extraction or chemical synthesis often struggles to meet current needs in terms of yield, titre, cost, and environmental impact. Benefiting from the rapid development of metabolic engineering and synthetic biology, microbial production of natural products from inexpensive and renewable sources provides a feasible solution for sustainable supply. This review outlines the biological activities of simple phenylpropanoids, compares their biosynthetic pathways in different species (plants, bacteria, and fungi), and summarises key research on the microbial production of simple phenylpropanoids over the last decade, with a focus on engineering strategies that seem to hold most potential for further development. Moreover, constructive solutions to the current challenges and future perspectives for industrial production of phenylpropanoids are presented.
1. Introduction
Simple phenylpropanoids are a group of secondary metabolites comprising a phenyl ring linked with a three-carbon side chain (C6–C3). In plants, they are primarily derived from the aromatic amino acid L-phenylalanine (L-Phe).1 These compounds can be classified into several subcategories associated with changes in the substituent on the benzene ring and the position of the propenyl double bond, such as phenylpropanoic acids (cinnamic and hydroxycinnamic acids), phenylpropanoic aldehydes, phenylpropanols, phenylpropene and simple coumarins.2 By serving as biogenetic precursors of various complex phenylpropanoids and other downstream metabolites, simple phenylpropanoids are essential for structural support, pigmentation, defence, and signalling throughout the plant kingdom.3,4 They also show a wide range of biological activities and are useful as raw materials applicable to high value-added chemical products, which have received considerable attention from agriculture, cosmetics, biofuel, biomaterials, and pharmaceutical industries.5,6Plants synthesize approximately 10 gigatons of phenylpropanoid molecules each year, which constitute approximately 20% of the total carbon in the terrestrial biosphere.7 Simple phenylpropanoids are mainly obtained from natural raw materials, particularly from millions of tons of agricultural wastes and forest litter produced per annum.8 Access to a sustainable supply of simple phenylpropanoids is restricted by a variety of factors, including sluggish growth and accumulation, variable synthesis fluctuations caused by climatic and environmental changes, challenging extraction and purification methods, as well as time-consuming and contaminating procedures.9,10 Development of more nature-friendly production methods has long been the subject of research owing to the intensifying contradiction between the increasing demand for simple phenylpropanoids and the urgent need for ecological conservation. Compared to plant extraction and chemical synthesis, microbial production is a promising alternative, not only for its much shorter production periods but also for being safer, low-cost, and environmentally friendly. Recent advances in the design-build-test-learn cycle associated with metabolic engineering, synthetic biology, systems biology, bioinformatics, and other advanced technologies have accelerated the development of microbial cell factories, resulting in the accumulation of simple phenylpropanoids from inexpensive and renewable sources.11–13 However, despite the tremendous progress made, several urgent challenges remain in advancing microbial engineering as a general approach for the biosynthesis of simple phenylpropanoids, including (1) non-specificity and inefficient cofactor supply of enzymes, (2) unbalanced metabolic flux between glycolysis and pentose phosphate pathway (PPP), (3) low cell performance, and (4) metabolic promiscuity in single cells.
In this review, we first summarize the extensive biological activities of simple phenylpropanoids, and emphasize the importance in food, cosmetic, nutraceutical, chemical, and, especially, the pharmaceutical industries. In addition, we outline biosynthetic pathways of simple phenylpropanoids in different species (plants, bacteria, and fungi), which provides inspiration for route design and optimization of microbial cell factories for heterologous synthesis. Next, we summarize effective engineering strategies for the microbial production of simple phenylpropanoids that are based on the reprogramming the metabolic flux toward aromatic amino acids (AAAs), and efficient microbial production of simple phenylpropanoids. Finally, we here discuss in detail the potential challenges for further improving titers of simple phenylpropanoids in microbial cell factories under the concept of the four-dimensional metabolic engineering. We also present some perspectives and constructive solutions to the current challenges.
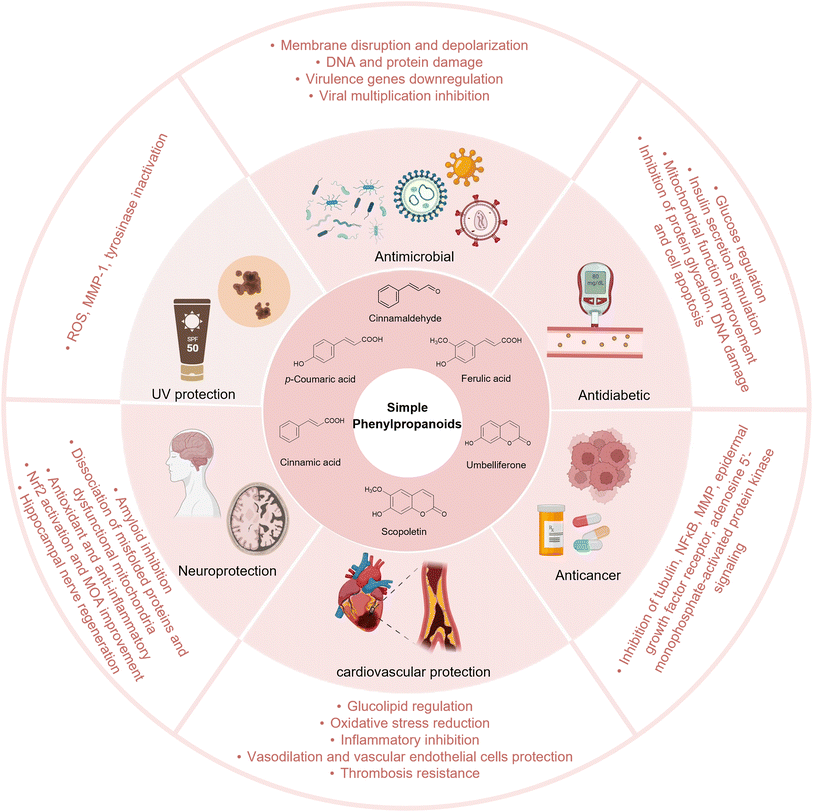
2. Biological activities
Simple phenylpropanoids are widely found in fruits, vegetables, cereals, coffee, tea, and some traditional herbs.14–16 They possess various biological activities, including antioxidant, anti-inflammatory, antimicrobial, antidiabetic, neuroprotective, and anticancer properties.17–20 In addition to their pharmacological activities, simple phenylpropanoids are core precursors to many complex natural products, such as flavonoids, lignans, and polyphenols. Therefore, simple phenylpropanoids play a significant role in food, cosmetic, nutraceutical, chemical, and, especially, the pharmaceutical industries (Fig. 1).2.1. Bioactivities of simple phenylpropanoids
2.2. Bioactivities of complex phenylpropanoids
In addition to serving as versatile phytochemicals, simple phenylpropanoids are key precursors of valuable complex natural products. The lignan compound podophyllotoxin has significant antitumor activity and is the raw material used in synthesis of etoposide, a clinically important chemotherapy drug.59 Silybin, a flavonolignan, exerts a therapeutic effect on hepatic disease.60 Icaritin is extracted from the Chinese Herba Epimedii and has been developed as a clinical drug for the treatment of liver cancer in China. Icaritin shows anti-inflammatory and immunomodulatory effects.61 Salvianolic acid B is the major bioactive water-soluble polyphenolic acid of Salvia miltiorrhiza, exhibits anti-inflammatory, anticancer, and cardioprotective activities, and has been clinically utilised to treat cardio- and cerebrovascular disorders.62,63Current efforts have developed novel forms such as nanoparticles to enhance bioavailability, and many artificial derivatives based on simple phenylpropanoid scaffolds have been synthesised to further expand applications.64
3. Biosynthetic pathways
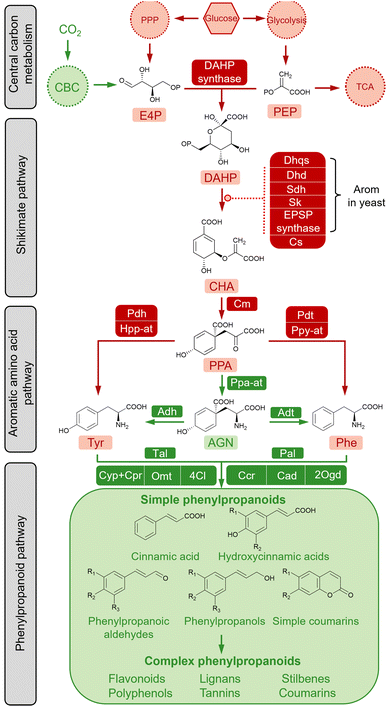
The diversity and conservatism of simple phenylpropanoids are the result of efficient modification and amplification towards the fundamental “C6–C3” structure through an orchestrated cascade of enzymes, including oxygenases, reductases, ligases and transferases.65 The biosynthesis of simple phenylpropanoids are derived from the aromatic amino acids (AAAs) L-Phe and L-tyrosine (L-Tyr) in plants (Fig. 2). As nodes connecting primary and secondary metabolism, L-Phe and L-Tyr are synthesised from chorismate (CHA) via the AAA pathway. The condensation of two core precursors, phosphoenolpyruvate (PEP), derived from glycolysis, and erythrose 4-phosphate (E4P), derived from the pentose phosphate pathway (PPP), leads to a metabolic flux into the shikimate pathway for CHA synthesis. Therefore, the biosynthetic pathways of simple phenylpropanoids can be divided into primary metabolism and plant-specific phenylpropanoid metabolism. Except for plant-specific CO2 fixation (shown in green in Fig. 2), the AAA biosynthetic pathway is relatively conserved in plants and microorganisms (shown in red in Fig. 2). Biosynthetic pathways of simple phenylpropanoids and their derived complex phenylpropanoids usually exist in plants.Plants absorb carbon dioxide to produce sugar, whereas glucose is the most direct carbon source for cellular biomass in bacteria and fungi. Central carbon metabolism converts glucose to PEP and E4P via glycolysis and nonoxidative steps in the PPP, respectively. Notably, PPP is related to the redox cofactor NADPH, which is critical for several downstream enzymes and the normal cellular energy metabolism.66,67 The pathway connecting central carbon metabolism and the AAA network is the shikimate pathway, which comprises seven enzymatic reactions and is ubiquitous in plants, fungi and bacteria.68 As the first committed step, studies have shown that the aldol condensation of PEP and E4P to produce 3-deoxy-D-arabino-heptulosonate 7-phosphate (DAHP) in a reaction catalysed by DAHP synthase is a rate limiting reaction.69–71
The following steps to generate the final product of the shikimate pathway, chorismate (CHA), are typically supported by individual monofunctional enzymes in plants and E. coli, including 3-dehydroquinate synthase (Dhps), 3-dehydroquinate dehydratase (Dqd), shikimate dehydrogenase (Sdh), shikimate kinase (Sk), 5-enolpyruvate-shikimate-3-phosphate synthase (Epsp synthase) and chorismate synthase (CS), whereas in Saccharomyces cerevisiae, a penta-functional protein (Arom) can directly catalyse the reaction of DAHP to 5-enolpyruvylshikimate-3-phosphate (EPSP).72,73 As a key branching point of the primary metabolic pathway to make aromatic amino acids, CHA firstly forms prephenate (PPA) in a reaction catalysed by chorismate mutase (Cm). The subsequent conversion to Phe and Tyr occurs via two routes in plants. One is conserved in both plants and microorganisms, where PPA catalyses the synthesis of L-Phe by prephenate dehydratase (Pdt) and phenylpyruvate aminotransferase (Ppy-at) or L-Tyr by prephenate dehydrogenase (Pdh) and 4-hydroxyphenylpyruvate aminotransferase (Hpp-at). PPA is converted to L-arogenate (AGN) by prephenate aminotransferase (Ppa-at), which is then converted in reactions catalysed by arogenate dehydratase (Adt) or arogenate dehydrogenase (Adh) to L-Phe or L-Tyr, respectively.74,75
The biosynthetic pathway producing simple phenylpropanoids that occurs in plants has been heterologously established in microorganisms—the specific pathways involved are described in the following section. The conversion of L-Phe to t-CA is catalysed by phenylalanine ammonia-lyase (Pal). Studies have demonstrated that the broad substrate spectrum of Pal includes L-Phe and L-Tyr, and that the reverse reaction to L-Phe can also be catalysed by Pal in the presence of ammonia.76t-CA is modified by the hydroxylation and methylation of the aromatic ring, giving rise to hydroxycinnamic acids. Among these, p-HCA can also be directly formed from the deamination of L-Tyr by the bifunctional Phe/Tyr ammonia-lyase (PTal) enzyme from monocot grasses of the Poaceae family, or from some bacterial and fungal species.77 The enzyme 4-coumarate-CoA ligase (4Cl) catalyses the conversion of phenylpropanoic acids to the coenzyme A (CoA) thioesters, which are then converted to the corresponding aldehydes by cinnamoyl-CoA reductase (Ccr), and subsequently to phenylpropanols by cinnamyl alcohol dehydrogenase (Cad), and simple coumarins by 2-oxoglutarate-dependent dioxygenase (2Ogd). Furthermore, simple phenylpropanoids are considered basic building blocks for downstream decorations to produce large quantities of secondary metabolites, such as flavonoids, stilbenes, lignans, polyphenols, and condensed tannins.65 These complex phenylpropanoids and their derivatives have various biological and commercial values and are being intensively evaluated.78
4. Microbial production
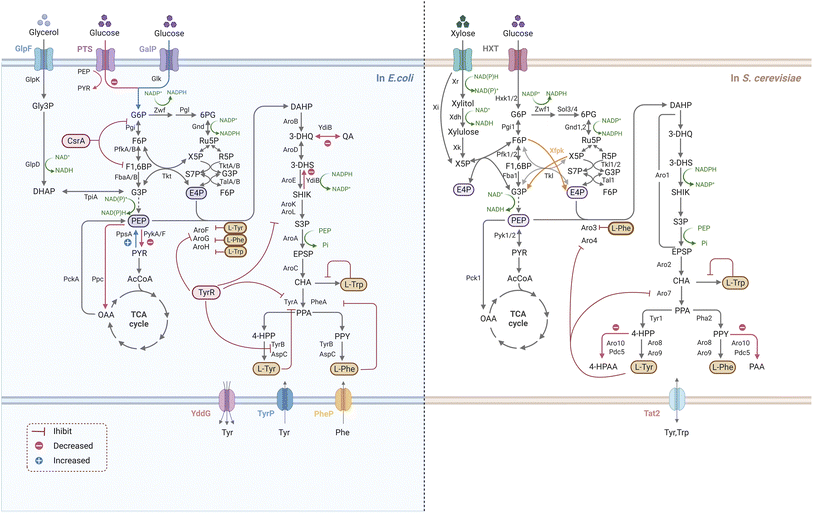
As the compounds connecting the microbial innate pathway and the plant allogenic pathway, L-Phe and L-Tyr obtain carbon fluxes from the central carbon metabolic and shikimate pathways for the synthesis of simple phenylpropanoids (Fig. 2). Reprogramming the primary metabolic flux to AAAs (L-Phe and L-Tyr) is the basis for the high-level synthesis of simple phenylpropanoids. Therefore, the construction of a microbial cell factory for the efficient synthesis of simple phenylpropanoids requires two steps: construction of an AAA high-producing chassis (Fig. 3) and introduction and optimization of simple phenylpropanoid pathways (Fig. 4).4.1. Efficient microbial production of AAAs
Multilevel metabolic engineering strategies have been studied to overproduce AAAs (L-Phe and L-Tyr), such as rewiring central carbon metabolism, relieving negative regulation, eliminating by-product formation, and engineering transport processes (Fig. 3). To rewire the central carbon flux towards the AAA pathway, an improved supply of PEP and E4P is required. The PPP is the exclusive pathway for glucose conversion to E4P, and its optimization is crucial for E4P supply. Since S. cerevisiae tends to quickly introduce metabolic flux into glycolysis for ethanol fermentation under anaerobic conditions and limits PPP, the deficiency of E4P is a major problem.79 In addition, E4P has been reported to be the primary limiting substrate for AAA biosynthesis in some bacteria, such as E. coli, B. subtilis, and Corynebacterium glutamicum.80–82 In S. cerevisiae, the optimization strategy for PPP depends on cell culture conditions because the upstream (Zwf and Gnd) and downstream (Tkl and Tal) enzymes of PPP are rate-limiting steps for E4P synthesis under high- and low-glucose concentrations, respectively.83 Therefore, different strategies should be used for PPP optimization to accommodate the use of high- and low-glucose-induced promoters.83–85 A novel PHK pathway consisting of phosphoketolase (Xfpk) can split xylose 5-phosphate (X5P) and/or fructose 6-phosphate (F6P) into acetyl-phosphate and glyceraldehyde-3-phosphate (G3P)/E4P, which is critical for rewiring the carbon metabolism of glycolysis into E4P.86 Therefore, simultaneous expression of Xfpk, Tal, and Tkl can significantly increase the metabolic flux towards E4P in S. cerevisiae.83 Although S. cerevisiae is not able to use xylose as a sole carbon source, it is clear that metabolic flux enters the PPP faster for E4P synthesis when xylose is used, which may be useful in further improving the E4P supply in yeast.87 In contrast with E4P, carbon flow towards PEP is usually sufficient because of its important position in glycolysis. Construction of a non-PTS route in E. coli has been proven to avoid PEP loss for glucose transport, (>50%), thereby supporting the synthesis of downstream AAA.88 The availability of PEP and E4P can also be improved by adjusting the activity of precursor pathway enzymes, such as by deletion of pyruvate kinases (encoded by PYKA and PYKF), or by over-expression of PEP synthase (encoded by PPSA), transketolase (encoded by TKTA) and/or transaldolase (encoded by TALB).89–91 It is reasonable that the carbon flux between PEP and E4P is significantly different, thus it is essential to balance the availability of E4P and PEP (increase E4P and decrease PEP) to maintain high productivity. Promoter engineering by fine-tuning the strength of glycolysis and PEP alleviated this limitation in S. cerevisiae.92,93Since bypass inevitably diverts the flow of the target pathway, it is necessary to control the pathway by weakening its branches to facilitate product synthesis. In E. coli, the bifunctional enzyme (YdiB) was replaced by the monofunctional enzyme (AroE) to reduce the by-product formation of quinate in the shikimate pathway.94 In S. cerevisiae, increased flux in the AAA biosynthetic pathway results in the accumulation of undesired fusel alcohol/acids via the Ehrlich pathway, which can be relieved by a double knock-out of phenylpyruvate decarboxylase (Aro10) and pyruvate decarboxylase (Pdc5).95 Interestingly, a recent report has found that the deletion of a novel gene HTZ1 significantly increased the tyrosine-producing capacity of yeast cells, although the underlying mechanism remains to be elaborated.96 Primary metabolism is strictly regulated by microorganisms, including yeasts. At the enzyme activity level, metabolic flux is controlled mainly by AAA-mediated feedback inhibition in the first and post-chorismate steps in the above-mentioned biosynthetic pathways. By protein structure analysis with site directed mutagenesis, the feedback-resistant (fbr) variants such as aro4K229L and aro7G141S have been widely utilized in S. cerevisiae to eliminate the allosteric regulation for the enhancement of the corresponding AAAs and derived metabolites.71,97,98CSRA, encoding the carbon-storage regulator of E. coli, regulates enzymes in glycolysis, and over 2- fold increase in yield of L-Phe were achieved when CsrA was disrupted.99 TyrR is the tyrosine repressor that negatively regulate several genes transcription of Tyr-producing pathway in E. coli, and inactivation of TyrR led to an increase in the L-Tyr biosynthesis.100,101 Transport engineering contributes to strain development by addressing substrate uptake and product output, as well as loss of intermediates. The tyrosine-specific transporters TyrP and TyrR were mutated in E. coli overexpressing aroGΔfbr, aroL and tyrC, leading to a maximum L-Tyr titre of 43.14 g L−1 by fed-batch fermentation.102
Notably, the regulation of cell growth and highly tailored production should be carried out in an optimal and sustainable manner. A synthetic RNA (sRNA) strategy was employed in E. coli to fine-tune target gene expression, which could coordinate the flux balance between L-Tyr production and biomass accumulation.103 Adaptive laboratory evolution could also be useful as a way of enhancing the robustness of engineered strains. A Pseudomonas putida S12 mutant strain highly accumulating L-Phe was successfully screened by a combination of random mutagenesis and selection of toxic analogs.104 Recently, with the aid of biosensors as a high-throughput screening tool, recombinant strains with the desired properties for enhanced AAAs are likely to be identified in high efficiency.105,106 Researchers have explored the potential of non-conventional microbial chassis to generate AAAs, such as Yarrowia lipolytica and C. glutamicum, which have been an attractive platform optimized for aromatic compounds production.107,108
4.2. Efficient microbial production of simple phenylpropanoids
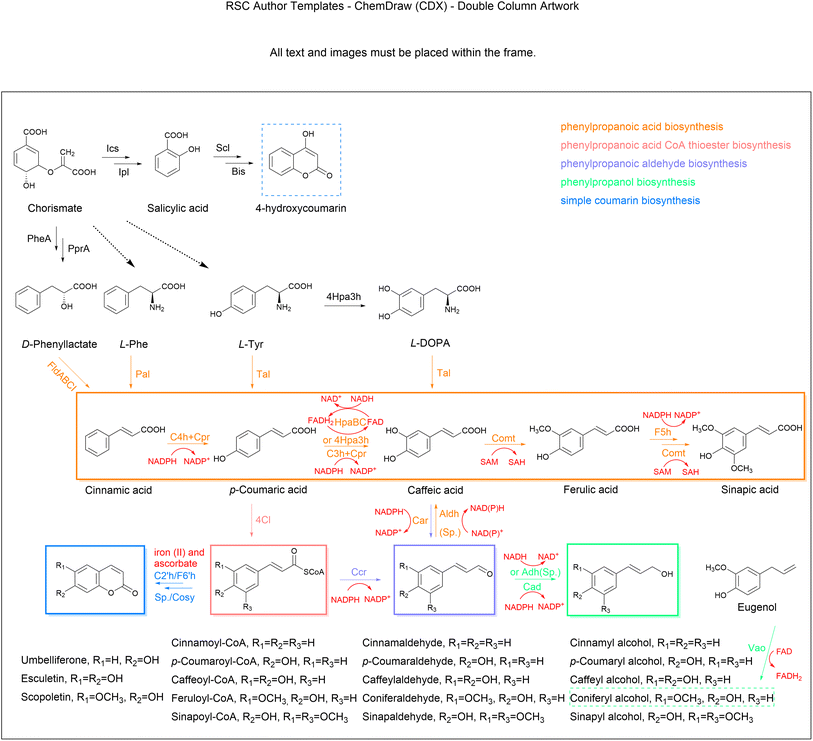
4.2.1.1. Cinnamic acid. Cinnamic acid belongs to a class of auxins that regulate cell growth and differentiation. Owing to its broad applicability, the microbial production of cinnamic acid, which is a relatively facile and green technology, has been actively explored over the last few decades. Cinnamic acid is the non-oxidative deamination product of L-Phe in a reaction catalysed by Pal and mostly occurs in the trans configuration (t-CA) (Fig. 4). Pal is the starting and limiting enzyme in the simple phenylpropanoids biosynthetic pathway, hence much effort has been made in identifying high-efficiency alternative enzymes for functional expression in heterologous microbes.109,110 The production titre of t-CA reached 6.9 g L−1 by heterologous expression of an efficient SmPal from Streptomyces maritimus in an engineered L-Phe-overproducing E. coli, together with cultivation optimization and casamino acid supplementation in a 2 litre bioreactor.111 In order to further optimize the catalytic function of SmPal, the combination of a SmPal-based whole-cell biocatalyst in C. glutamicum and a crossflow membrane-based cell recycling system was designed to produce t-CA from L-Phe with a yield of 75% (0.75 mol mol−1).112 An alternative pathway harbouring a phenyllactate dehydratase encoded by FLDABCI genes from Clostridium sporogenes was developed for additional t-CA production from D-phenyllactate (Fig. 4), which exceeded the Pal-dependent pathway to achieve an 11-fold increase in t-CA yield.113 Most recently, a high-density culture system of the cyanobacterium Synechocystis sp. PCC 6803 successfully synthesized approximately 0.8 g L−1t-CA from CO2 as the carbon source,114 which indicates that exploiting the primary metabolism of cyanobacteria has great potential for the green and sustainable production of phenylpropanoids. As the cytotoxicity of t-CA impairs cell proliferation, researchers have found that near-neutral culture pH and alcoholic carbon source might be useful to improve the cellular tolerance to t-CA inhibition.115
4.2.1.2. Coumaric acid. Coumaric acid, also known as hydroxycinnamic acid, naturally exists as three isomers (ortho-, meta-, and para-) with different substitution positions for the active hydroxyl group on the benzene ring. Among these, para-coumaric acid (p-HCA) occurs more often in nature.116 With advances in metabolic engineering, microbial production has become a promising alternative for obtaining large-scale yields of p-HCA. Starting with aromatic amino acids, L-Tyr can be directly converted to p-HCA by Tal, which is typically found in bacteria. Alternatively, the deamination product t-CA of L-Phe can be further hydroxylated to p-HCA by membrane bound cinnamate-4-hydroxylase (C4h) in plants.117 The Tal-dependent one-step biosynthesis is more convenient and economical; also, the Pal-C4h pathway requires plant-derived Cpr and cofactor NADPH to form the electron transport chain to complete the hydroxylation reaction (Fig. 4). Therefore, the one-step reaction catalysed by Tal is favoured for application in p-HCA production from microbial cell factories.118–120 However, this conversion is often limited by the poor activity of the heterologous Tal enzyme and the competitive inhibition of p-HCA on Tal and Pal. Therefore, it is necessary to identify novel enzymes with higher and more stable activities from different sources. The substrate specificity and activity of enzymes can be altered by rationally designing active amino acid residues. By the combination of directed evolution and high-throughput screening, a Tal variant from Rhodotorula glutinis with higher selectivity for L-Tyr and superior catalytic efficiency was screened, producing a p-HCA titre that was 2.2-fold higher than that of the control strain.121 Systematic bioinformatics analysis and enzyme characterization have been applied to identify Tals most suitable for p-HCA production in E. coli and yeast.122,123 Interestingly, when the FjTal protein was anchored to the yeast vacuole, p-HCA production was significantly enhanced.124 Although C4h as a membrane-bound Cyp is not amenable to expression in E. coli, a novel C4h from Lycoris aurea was identified and functionally expressed in E. coli, and when combined with the optimization of NADPH regeneration and Cpr expression enabled a p-HCA titre of 25.62 mg L−1.125 In order to greatly enlarge the capacity of yeast in producing p-HCA, carbon metabolic allocation is optimized towards aromatic amino acid biosynthesis, which, together with the microbial Tal and introduction of a plant Pal-C4h, led to a high-level production of p-HCA (12.5 g L−1) in fed-batch fermentation in S. cerevisiae.126 To eliminate the Crabtree effect, xylose as the carbon source for p-HCA production was investigated in S. cerevisiae and finally achieved a 45-fold increase in p-HCA production compared to the use of glucose as the carbon source under the same conditions. In recent years, the de novo synthesis of p-HCA in Synechocystis sp. PCC 6803 has been achieved. Using the fixation of CO2 by solar energy as the carbon source, the highest total titre was up to 0.4 g L−1 in a high-density cultivation system.127,128 To eliminate the genetic and production instability of strains using episomal plasmid expression in the fermentation, a high pHCA-yielding S. cerevisiae was built through POT1-mediated delta integration, after which the titre and gene copy number remained stable for more than one hundred generations.129 Transporters are responsible for the efflux of small product molecules, and have showed significance in strain improvement. The deletion of a specific transporter (TAT1), which is responsible for transport of L-Tyr and L-Trp, led to a 50% increase in p-HCA titre.130 Upregulation of the aromatic acid transporter ESBP6 promoted increases in the p-HCA titre, as well as improvements in the yeast tolerance to toxicity.131 Although the microbial production of p-HCA has already achieved on a g L−1 scale, microbial cell factories still need to be further optimized for high-level synthesis, such as by targeting product degradation and inhibition, cofactor imbalance, and transcriptional regulation.132,133
4.2.1.3. Caffeic acid. CaA is a phytonutrient frequently found in plants, including vegetables. The biosynthesis of CaA involves the ortho-hydroxylation of p-HCA through the Cyp enzyme p-coumarate 3-hydroxylase (C3h) (Fig. 4), which is traditionally difficult to perform in bacteria.134 Therefore, identifying different C3h isozymes from other organisms is crucial for the synthesis of CaA in engineered cells. A microbial C3h encoded by SAM5 from Saccharothrix espanaensis and a site-directed mutant of CYP199A2 from Rhodopseudomonas palustris were shown to have activity when expressed in bacteria to synthesise CaA from p-HCA.135,136 The de novo biosynthesis of CaA in E. coli was also achieved by expressing endogenous 4-hydroxyphenylacetic acid 3-hydroxylase (4Hpa3h) that can hydroxylate both p-HCA and L-Tyr to generate CaA and L-3,4-dihydroxyphenylalanine (L-DOPA), respectively, while L-Dopa was further converted to CaA by Tal from Rhodobacter capsulatus.91 An alternative pathway catalysed by bacteria-coupled enzymes HpaB (FAD-dependent 4-hydroxyphenylacetate-3-monooxygenase) and HpaC (NADH-flavin oxidoreductase) has been shown to be the most preferred strategy for CaA synthesis in both bacterial and fungal systems.137,138 In this pathway, the direct electron donor is FADH2, rather than NADPH, and FADH2-dependent enzymes are usually more adapted in microorganisms.139 The yield of CaA also depends on the compatibility of HpaB and HpaC in cell factories. Through orthogonal assays, the combination of HpaB from Pseudomonas aeruginosa and HpaC from Salmonella enterica was regarded as the best.140 By expressing this enzyme pair, combined with the cofactor optimization strategy, the yield of CaA in S. cerevisiae reached 5.5 g L−1.83 This strategy using suitable cofactor engineering provides fundamental insights for future research in other natural product biosynthesis. Apart from enzyme screening and cofactor engineering, control of transport proteins has also been proven to be an effective strategy for reducing cytotoxicity and promoting CaA synthesis. By overexpressing the putative sugar ABC transporter permease (YcjP), which was identified through transcriptome data mining, the production of CaA was further enhanced to about 7.9 g L−1 in E. coli with the integration of other optimized factors. This is the highest known titre so far.141 Recently, the multi-copy integration expression strategies based on delta sites were applied to integrate the genes of the CaA pathway into yeast and resulted in an increase of CaA production by 50 times compared to that produced by the initial multi-copy-plasmid expression.142 Some other carbon sources and the whole-cell biocatalyst strategy have also been successfully used for CaA synthesis.143,144 By introducing the xylose assimilation pathway into Candida glycerinogenes, the advantage of using mixed sugars as carbon sources showed that the optimized strain eventually obtained 1.2-fold higher CaA than that using glucose.145 In addition, the generation of CaA can proceed in four steps, from L-Tyr through p-HCA, coumaroyl-CoA and caffeoyl-CoA, as CoA thioesterase can convert caffeoyl-CoA to CaA, although this pathway is inefficient.146,147
4.2.1.4. Ferulic acid. FA (p-hydroxy-3-methoxycinnamic acid), a phytomolecule crosslinked with lignin and hemicellulose in plant cell walls, is abundant in certain cereal raw materials and medicinal herbs.148 Caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase (Comt) catalyses the methylation of the 3-hydroxyl group of CaA to generate FA, which can be further converted to sinapic acid (Fig. 4). SA has multiple pharmaceutical applications,149 but limited data are available on its microbial production. Most efforts have been devoted to constructing FA synthetic pathways in microbial hosts to produce FA using glucose or L-Tyr as substrates.150,151 For example, a recombinant E. coli harbouring plasmids with the pathway genes (TAL, C3H and COMT) produced 257.3 mg L−1 FA from L-Tyr.152 In another example, after the optimization of gene expression by changes to the promoter strength and copy number, coupled with enhanced NADPH levels to improve the conversion towards CaA, and overexpression of a methionine kinase, 212 mg L−1 of FA was produced in shake flask cultures.153 More recently, an engineered yeast achieved a yield of 3.8 g L−1 of FA in a fed-batch fermentation through cofactor engineering of that accelerated the methyl cycle and SAM regeneration.83 To address the insufficient supply of HpaBC-dependent cofactor FADH2, a NAD(P)H-flavin reductase (Fre) was introduced to activate FADH2 regeneration, which promoted an 8.1-fold increase in efficiency of hydroxylation. Combined with the efforts on a L-Tyr overproducer and SAM reactivation, the total titre of FA reached 5.09 g L−1 in E. coli under the fed-batch condition.154 FA and SA can also be synthesized from the oxidation of coniferyl aldehyde and sinapoyl aldehyde, respectively, by the corresponding aldehyde dehydrogenase (Aldh), which indirectly detoxify the reactive aldehydes in cellular metabolic processes.155,156 Alternatively, the biotransformation of eugenol to FA was also established in a recombinant S. cerevisiae by expressing vanillyl-alcohol oxidase (PsVao) from Penicillium simplicissimum, and 16.9 g L−1 of FA was produced from eugenol feeding in a fermentor.157 Of special interest, the Vao family of enzymes involve many reactions with phenolic substrates and the FAD cofactor. Considering that eugenol is an inexpensive and readily available substrate, its bioconversion to FA is likely to expand to industrial levels. Notably, an alternative one-step production method based on microbial fermentation is of great interest, using feruloyl esterase to prepare FA from agro-industrial wastes (e.g., wheat bran and brewery spent grain) with microbial fermentation.158–160 Feruloyl esterase is expected to be a catalyst for the production of biofuels from biomass.
The biotechnological production of natural phenylpropanols has been achieved by reconstructing the plant monolignol pathway in microbial hosts. Although the synthesis of cinnamyl alcohol and hydrocinnamyl alcohol has been achieved in S. cerevisiae and E. coli by expressing Cad or Ccr enzymes from different species,167–169 severe product inhibition is the key limiting factor in cinnamyl alcohol biosynthesis. Product inhibition was successfully removed using a dibutyl phthalate/water biphasic system, which constantly separated and concentrated cinnamyl alcohol synthesised by E. coli from t-CA into the organic phase.170 Coniferyl alcohol, a precursor of silybin and other natural pharmaceuticals, is the most abundant monolignol in plants. The co-expression of 4Cl, Ccr, and Cad from Arabidopsis thaliana in a FA producer achieved de novo synthesis of coniferyl alcohol with titres of 187.7 mg L−1 and 201.1 mg L−1 in E. coli and S. cerevisiae through fed-batch fermentation, respectively.171,172 Moreover, it is noteworthy that these enzymes responsible for the monolignol biosynthesis are known to be promiscuous on the substrate specificity, which is double-edged for the production of phenylpropanols.173,174 A study applied a co-culture strategy to the engineered E. coli hosts, which minimized the effect of promiscuous HpaBC catalysing the side reaction of L-Tyr to produce L-Dopa, reaching 534 mg L−1 caffeyl alcohol and 124.9 mg L−1 coniferyl alcohol.137 Additionally, a study used the PsVao to convert eugenol to coniferyl alcohol in E. coli, together with catalase (Cta1) from S. cerevisiae to avoid over-oxidation of coniferyl alcohol, which reached the final coniferyl alcohol titre of 53.9 g L−1 in a 5 L bioreactor with a conversion rate of 86.72%.175 In another study, E. coli was demonstrated to synthesize non-natural phenylpropanols through feeding of different precursors, such as 5-bromoconiferyl alcohol and 2-nitroconiferyl alcohol.176
To prevent the degradation of 4Cl-producing thioester intermediates, a predicted acyl-CoA thioesterase (YbgC) was deleted for higher production of esculetin and umbelliferone from glucose.183 A systematic study that overcame the limitation of 4Cl through protein engineering improved the supply of L-Tyr through metabolic flow remodelling and optimized fermentation conditions, obtaining 356.59 mg L−1 umbelliferone from L-Tyr.184 To overcome the inefficient spontaneous reactions, researchers established a novel artificial pathway condensing malonyl-CoA and salicylic acid by a biphenyl synthase (Bis) that was introduced into E. coli for the de novo synthesis of 4-hydroxycoumarin from glycerol. The resulting titre was 483.1 mg L−1 in 24 h.185 Furthermore, the production was enhanced to 935 mg L−1 in shake flasks by alleviating the thioesterase-mediated degradation of salicoyl-CoA.186 Nowadays, the use of lignin hydrolysate to produce valuable chemicals with benzene rings has attracted much interest in biotransformation technology. An engineered budding yeast expressing necessary enzymes to generate scopoletin from lignin hydrolysate was recently reported.187 The scopoletin production reached 4.79 mg L−1, suggesting that this approach may offer new opportunities for improved biosynthesis of coumarins from renewable sources.
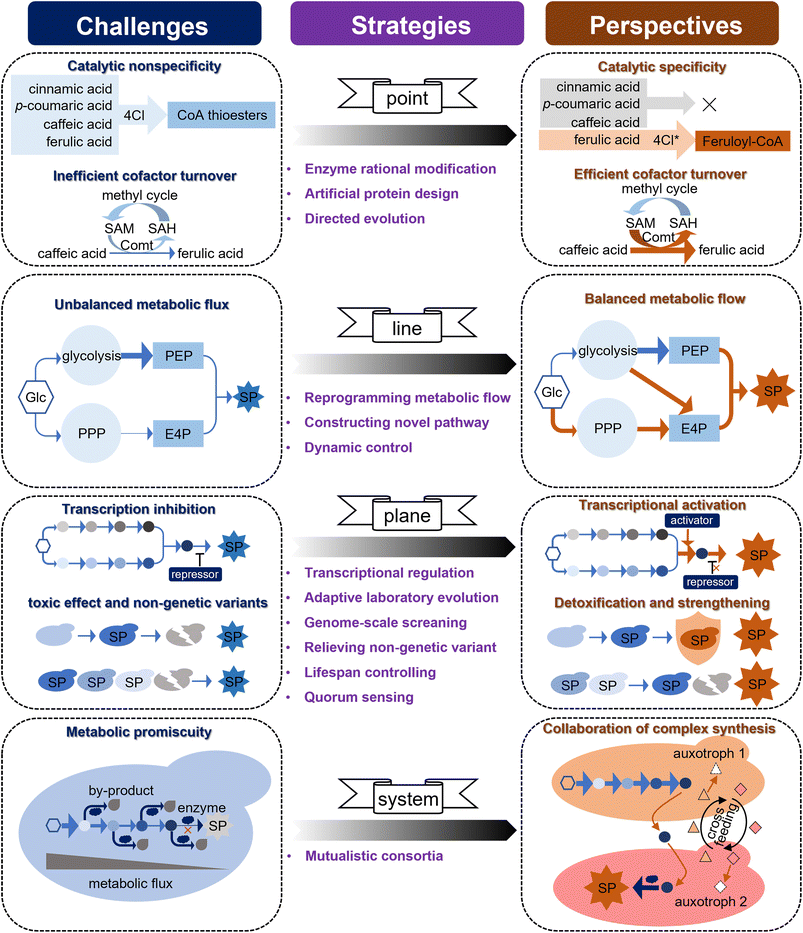
5. Discussion and future perspectives
Driven by the important applications of simple phenylpropanoids, either because of their wide bioactivity or as precursors for the synthesis of complex natural product molecules, great progress has been achieved in the microbial production of simple phenylpropanoids (Table S1 provided as an attachment).† Although many simple phenylpropanoids have been synthesised at the g L−1 level in microorganisms with considerable engineering endeavours over the past 20 years, their titre, yield, and productivity (TYP) still require improvement to achieve industrial production. Some challenges that limit the high-level synthesis of simple phenylpropanoids must be addressed. Based on the three-dimensional metabolic engineering strategy proposed in the previous study,11 here we map these challenges to an upgraded four-dimensional metabolic engineering strategy (i.e., “point-line-plane-system”), and suggest a potential technical proposal to meet these challenges (Fig. 5).(i) The “Point” level: the unsatisfied enzyme activity and cofactor utilization.
Enzymes are the most basic elements that fundamentally determine the synthetic efficiency in a cell factory through specific activities and cofactor supply. The catalytic promiscuity and low cofactor utilisation of enzymes in the simple phenylpropanoid pathway are key issues hindering synthetic efficiency. 4Cl is a rate-limiting enzyme with a broad substrate spectrum that generates CoA thioester intermediates from phenylpropanoic acids (Fig. 4), which may result in unexpected disruption of metabolic flux. For instance, when 4Cl is designed to react with FA, it preferentially reacts with upstream phenylpropanoic acids (i.e., t-CA, p-HCA, and CaA), thereby blocking the metabolic flux towards FA. Given that several 4CL protein crystal structures have been resolved (UniProt Q42524, Q9SMT7, and Q94M3), and there have been some advances in changing the 4Cl substrate preference.188 It is expected that specific unnatural variants of 4Cl can be designed through classical structural biology-based protein rational modification. The continued evolution of machine-learning-assisted models will provide feasible clues for the virtual screening and prediction of key enzymes.189,190 More importantly, the latest advances in deep learning-inspired language models may enable the de novo design of enzymes, which, in the case of ProGen, can be adapted to generate artificial protein sequences that are functionally identical to natural proteins.191 Comt is the key enzyme in FA synthesis, and its low turnover of the methyl donor SAM results in low catalytic efficiency for CaA. Although the conversion rate of CaA to FA was increased from 28% to 64% through the metabolic engineering strategy of expressing an efficient form of Comt and by optimising the methyl cycle,83 more than 36% of CaA still failed to synthesise FA, and the catalytic problem of Comt itself has not been solved. Biosensors are an ideal strategy for high-throughput screening of enzymes based on intuitive phenotypic changes. A FARON switch system derived from a phenolic acid decarboxylase regulator (aPadR) was designed to respond to FA in mammalian cells.192 In addition, a sensitive synchronous fluorometric method based on the oxidation of FA with Ce(IV) in a sulfuric acid medium was used to detect FA in vitro. Therefore, optimization of the FARON biosensor and the Ce(IV)-dependent sensor to improve the cofactor turnover of Comt through directed evolution is expected to further increase FA production.
(ii) The “Line” level: the imbalance of multi-pathway metabolic fluxes.
Simple phenylpropanoids are derived from two AAAs (L-Phe and L-Tyr) condensed from two endogenous intermediates, i.e., E4P from PPP and PEP from glycolysis. Therefore, an equal supply of E4P and PEP is advantageous for the synthesis of simple phenylpropanoids. Although glycolysis and PPP are the two fundamental routes for catabolizing glucose in most living cells, E4P has been reported to be the primary limiting substrate for simple phenylpropanoid biosynthesis in many microorganisms.80,193 To maintain the intracellular balance of NADH/NAD+ and a fast growth rate, S. cerevisiae tends to introduce metabolic flux into glycolysis for ethanol fermentation and limits PPP when consuming sugars under anaerobic conditions.79 Some attempts to control heterologous expression under glucose-limited conditions, stretching PPP fluxes and weakening glycolysis have successfully increased the E4P supply and simple phenylpropanoid production in S. cerevisiae.83 Although yeast central metabolism has been extensively rewired to improve the supply of E4P,126 it is challenging to balance metabolic flux because of the metabolic competition between PPP and glycolysis. Considering that S. cerevisiae can be modified from an ethanol-producing to an oil-producing yeast, this strong metabolic plasticity implies the possibility of further improvement in the metabolic flux towards E4P through metabolic reprogramming.194 Furthermore, a novel PHK pathway consisting of phosphoketolase (Xfpk) and phosphotransacetylase (Pta) was introduced to channel more carbon flux towards E4P from F6P derived from glycolysis.86 Metabolic flux analysis indicated that the availability of E4P and the unbalanced supply of E4P and PEP remain key issues. To further alleviate this limitation, the introduction of an efficient xylose pathway may be a viable option to improve E4P supply.195 In addition, it may be possible to construct a self-adjusting system196,197 or optogenetic regulation198 to allow the dynamic control of metabolic flux towards glycolysis and PPP.
(iii) The “plane” level: the weakness of cell performance.
Even for well-studied microorganisms such as E. coli and S. cerevisiae, it is often not easy to accurately understand and correct the negative effects of metabolic engineering on whole cells. These negative effects may be caused by transcriptional regulation, accumulation of toxic products, and unknown mechanisms. Ric1 is a transcriptional repressor of multiple genes in the aromatic amino acid biosynthetic pathway in S. cerevisiae, and inhibition of Ric1 has been shown to increase the yield of shikimic acid, a precursor of simple phenylpropanoids.199,200 Although the identified targets can be used to optimize the synthesis levels, most targets for the global optimization of microbial synthesis performance are unknown. Aromatic compounds are generally toxic to microorganisms, which makes their high-level production in microbial hosts challenging. Adaptive laboratory evolution is a powerful tool used in the improvement of microbial cell factories at the entire cell level (the “plane” level), which emphasizes the importance of “collaboration” between scientists' deliberate choices and microorganisms' initiative to achieve design goals, and compensates for the lack of comprehensive understanding of host strains. Coordination between adaptive laboratory evolution and whole-genome sequencing revealed that Esbp6 is an important transporter for the secretion of p-HCA and tolerance to aromatic acids, which can be used to optimize the production of simple phenylpropanoids.131 Inspired by this, we believe that evaluating the growth rate on a medium with high concentrations of aromatic acids in a genome-scale collection of S. cerevisiae gene-deletion strains will uncover novel strategies for improving aromatic acid tolerance.
During microbial fermentation, although the genetic information of all individual cells in a microbial population is identical, their contributions to compound synthesis and single-cell biosynthetic performance may vary greatly (up to a 10-fold difference) owing to nongenetic cell-to-cell variation, which could have a significant impact on group performance.201,202 Nongenetic cell-to-cell variations may be due to different epigenetic modifications and variation in the regulation of expression due to differences in developmental history (uneven cell division and different parent cells) or exposure to environmental factors (differences in the concentrations of various substances in the local medium). Understanding the key mechanisms affecting nongenetic variations requires long-term efforts that do not provide reliable short-term engineering strategies. In one study, a minority (15%) of the total cell population produced more than half of the total free fatty acids, and the majority of cells performed very weakly in the fermentation of a fatty acid-producing E. coli. A quality control (PopQC) system was constructed to continuously select and kill low-performing nongenetic strains and maintain high-performing nongenetic variants for production.203 Since such genetic circuits for screening often rely on specific regulatory elements, biosensors that respond to simple phenylpropanoid concentrations should first be developed. Although this strategy alleviates the problems caused by nongenetic variants to some extent, it may inadvertently kill young cells with synthetic potential. Conversely, regulating cellular processes such as autophagy, apoptosis, and replication can extend the lifespan and the effective time available for product synthesis, thereby increasing the yield.204–206 Quorum sensing is a widespread bacterial mechanism for cell-to-cell communication that synchronises gene expression and has been successfully implemented to improve the synthetic performance of 4-hydroxycoumarin, flavonoids, and simple chemicals in bacteria.207–209 Although autoinducer-2 has been successfully used as a ‘universal signal’ for interspecies communication to improve CaA synthesis in S. cerevisiae,83,210 its mechanism and universality require further exploration. As an important mechanism of intercellular communication, an in-depth study of exosomes is expected to provide opportunities to improve the overall performance of cell factories.211
(iv) The “system” level:
As multicellular organisms, plants often complete the oriented synthesis of natural products through the division of labour and metabolite transport of multiple cells, tissues, organs, and organelles.212 The synthesis of aromatic acids involves the cooperation of multiple organelles, which is important for ensuring an optimal catalytic environment and avoiding metabolic promiscuity.213 Although compartmentalisation strategies can significantly improve synthesis efficiency by the modularized expression of heterologous pathways in different organelles, it sometimes comes at the expense of an optimal reaction environment for catalytic enzymes. For example, to avoid side reactions and waste of metabolic flux caused by the catalytic promiscuity of 4Cl, 4Cl can be considered as a node to separate the upstream and downstream pathways for the synthesis of ferulate CoA, a key precursor of many phenylpropanoids. However, both the upstream and downstream pathways need to be expressed in the cytosol to ensure the necessary reaction environment, such as NADPH and SAM cofactors. Therefore, building a stable and mutualistic microbial consortium system that cooperates with plant cells could reasonably confer wider metabolic capabilities and achieve more complex synthesis, which is difficult to achieve in a single cell. In addition, mutualistic consortia also endow microorganisms with higher synthetic titres through unique abilities, such as balancing metabolic flow, relieving metabolic burden, optimising resource utilisation, enriching the cellular environment and cofactors, and adapting to fluctuating environments. Considering these advantages, microbial consortium strategies have been extended to construct bacterial, yeast, and yeast systems for the synthesis of terpenoids,214 phenylpropanoids,215,216 and polyketones,217 and alkaloids.218 However, most are simply mixed cultures formed by changing the initial inoculation ratio,219 which makes it difficult to form a stable community, and this instability is further amplified in large-scale fermentation. Efforts to construct a mutualistic relationship for the members of microbial consortia to form stable mutualistic consortia is a viable approach to improve these strategies.216,220
Overall, based on the current gram-scale synthesis of simple phenylpropanoids in microorganisms, the integration of multidisciplinary technologies and tools is expected to further break the bottlenecks and increase production levels. These strategies will provide a framework for the synthesis of complex phenylpropanoids and other types of natural products.
6. Author contributions
Z. P. Zhu: conceptualisation, visualisation, writing – original draft, writing – review & editing. R. B. Chen: conceptualisation, founding acquisition, writing – original draft, writing – review & editing. L. Zhang: conceptualisation, supervision, founding acquisition, writing – review & editing.7. Conflicts of interest
There are no conflicts to declare.8. Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 82225047, 82170274 and 32000231), the National Key Research and Development Program of China (No. 2022YFC3501703), and the Shanghai Science and Technology Development Funds (No. 23QA1411400, China).9. References
- T. Vogt, Mol. Plant, 2010, 3, 2–20 CrossRef CAS.
- S. Wang, S. Alseekh, A. R. Fernie and J. Luo, Mol. Plant, 2019, 12, 899–919 CrossRef CAS PubMed.
- W. Biała and M. Jasiński, Front. Plant Sci., 2018, 9, 1610 CrossRef.
- N. Q. Dong and H. X. Lin, J. Integr. Plant Biol., 2021, 63, 180–209 CrossRef CAS.
- N. Redhu, A. Khatkar and K. K. Sharma, Crit. Rev. Food Sci. Nutr., 2020, 60, 2655–2675 CrossRef PubMed.
- R. J. Sharifi, M. N. Cruz, J. P. Lopez, E. P. Lopez, N. Harun, B. Yeskaliyeva, A. Beyatli, O. Sytar, S. Shaheen, F. Sharopov, Y. Taheri, A. O. Docea, D. Calina and W. C. Cho, Oxid. Med. Cell. Longevity, 2021, 2021, 6492346 Search PubMed.
- D. Peters, Adv. Biochem. Eng. Biotechnol., 2007, 105, 1–30 CrossRef CAS.
- A. Tilay, M. Bule, J. Kishenkumar and U. Annapure, J. Agric. Food Chem., 2008, 56, 7644–7648 CrossRef CAS PubMed.
- A. Nieter, S. Kelle, D. Linke and R. G. Berger, Bioresour. Technol., 2016, 220, 38–46 CrossRef CAS.
- C. J. Paddon, P. J. Westfall, D. J. Pitera, K. Benjamin, K. Fisher, D. McPhee, M. D. Leavell, A. Tai, A. Main, D. Eng, D. R. Polichuk, K. H. Teoh, D. W. Reed, T. Treynor, J. Lenihan, M. Fleck, S. Bajad, G. Dang, D. Dengrove, D. Diola, G. Dorin, K. W. Ellens, S. Fickes, J. Galazzo, S. P. Gaucher, T. Geistlinger, R. Henry, M. Hepp, T. Horning, T. Iqbal, H. Jiang, L. Kizer, B. Lieu, D. Melis, N. Moss, R. Regentin, S. Secrest, H. Tsuruta, R. Vazquez, L. F. Westblade, L. Xu, M. Yu, Y. Zhang, L. Zhao, J. Lievense, P. S. Covello, J. D. Keasling, K. K. Reiling, N. S. Renninger and J. D. Newman, Nature, 2013, 496, 528–532 CrossRef CAS.
- R. Chen, S. Yang, L. Zhang and Y. J. Zhou, iScience, 2020, 23, 100879 CrossRef CAS PubMed.
- A. Cravens, J. Payne and C. D. Smolke, Nat. Commun., 2019, 10, 2142 CrossRef PubMed.
- J. Nielsen and J. D. Keasling, Cell, 2016, 164, 1185–1197 CrossRef CAS PubMed.
- F. Destani, A. Cassano, A. Fazio, J.-P. Vincken and B. Gabriele, J. Food Eng., 2013, 117, 263–271 CrossRef CAS.
- M. G. S. Palmieri, L. T. Cruz, F. S. Bertges, H. M. Húngaro, L. R. Batista, S. S. da Silva, M. J. V. Fonseca, M. P. Rodarte, F. M. P. Vilela and M. d. P. H. do Amaral, Biocatal. Agric. Biotechnol., 2018, 16, 43–48 CrossRef.
- C. G. Schmidt, L. M. Gonçalves, L. Prietto, H. S. Hackbart and E. B. Furlong, Food Chem., 2014, 146, 371–377 CrossRef CAS.
- L. Zhao, X. Jin, Z. Xiong, H. Tang, H. Guo, G. Ye, D. Chen, S. Yang, Z. Yin and H. Fu, Int. J. Mol. Sci., 2022, 23, 11119 CrossRef CAS.
- G. Caruso, J. Godos, A. Privitera, G. Lanza, S. Castellano, A. Chillemi, O. Bruni, R. Ferri, F. Caraci and G. Grosso, Nutrients, 2022, 14, 819 CrossRef CAS PubMed.
- L. Liu, W. R. Hudgins, S. Shack, M. Q. Yin and D. Samid, Int. J. Cancer, 1995, 62, 345–350 CrossRef CAS.
- P. Khare, S. Jagtap, Y. Jain, R. K. Baboota, P. Mangal, R. K. Boparai, K. K. Bhutani, S. S. Sharma, L. S. Premkumar and K. K. Kondepudi, Biofactors, 2016, 42, 201–211 CrossRef CAS PubMed.
- T. R. E. Panel, D. Belsito, D. Bickers, M. Bruze, P. Calow, H. Greim, J. Hanifin, A. Rogers, J. Saurat and I. Sipes, Food Chem. Toxicol., 2007, 45, S1–S23 CrossRef.
- J. Jeon, J. Sung, H. Lee, Y. Kim, H. S. Jeong and J. Lee, J. Food Biochem., 2019, 43, e12701 CrossRef PubMed.
- M. Vaezi, J. Biomol. Struct. Dyn., 2023, 41, 4798–4810 CrossRef CAS.
- Y. Seo, S. Kim, Y. Boo, J. Baek, S. Lee and J. Koh, Clin. Exp. Dermatol., 2011, 36, 260–266 CrossRef CAS PubMed.
- A. M. Awad, P. Kumar, M. R. Ismail-Fitry, S. Jusoh, M. F. Ab Aziz and A. Q. Sazili, J. Food Process. Preserv., 2022, 46, e16796 CAS.
- L. Yue, D. Sun, I. M. Khan, X. Liu, Q. Jiang and W. Xia, Food Chem., 2020, 309, 125513 CrossRef CAS.
- I. b. Jantan, B. A. Karim Moharam, J. Santhanam and J. A. Jamal, Pharm. Biol., 2008, 46, 406–412 CrossRef.
- M. Friedman, J. Agric. Food Chem., 2017, 65, 10406–10423 CrossRef CAS PubMed.
- F. Khan, N. I. Bamunuarachchi, N. Tabassum and Y.-M. Kim, J. Agric. Food Chem., 2021, 69, 2979–3004 CrossRef CAS.
- A. T. Silva, C. M. Bento, A. C. Pena, L. M. Figueiredo, C. Prudêncio, L. Aguiar, T. Silva, R. Ferraz, M. S. Gomes, C. Teixeira and P. Gomes, Molecules, 2019, 25, 66 CrossRef.
- Y. Chen, Z. Li, P. Pan, Z. Lao, J. Xu, Z. Li, S. Zhan, X. Liu, Y. Wu and W. Wang, Antiviral Res., 2021, 192, 105117 CrossRef CAS.
- Y. Zheng, S. H. Ley and F. B. Hu, Nat. Rev. Endocrinol., 2018, 14, 88–98 CrossRef PubMed.
- S. Adisakwattana, Nutrients, 2017, 9, 163 CrossRef.
- W. Sompong, H. Cheng and S. Adisakwattana, J. Physiol. Biochem., 2017, 73, 121–131 CrossRef CAS.
- T. Jin and C. Chen, Food Chem. Toxicol., 2022, 163, 112892 CrossRef CAS PubMed.
- S. Kumar, B. K. Singh, A. K. Prasad, V. S. Parmar, S. Biswal and B. Ghosh, Eur. J. Pharmacol., 2013, 700, 32–41 CrossRef CAS.
- Y. Luo, K.-M. Qiu, X. Lu, K. Liu, J. Fu and H.-L. Zhu, Bioorg. Med. Chem., 2011, 19, 4730–4738 CrossRef CAS PubMed.
- C. Kopp, S. P. Singh, P. Regenhard, U. Müller, H. Sauerwein and M. Mielenz, Int. J. Mol. Sci., 2014, 15, 2906–2915 CrossRef PubMed.
- S. Li and S. Hu, Trop. J. Pharm. Res., 2020, 19, 957–963 CrossRef CAS.
- K. Pei, J. Ou, J. Huang and S. Ou, J. Sci. Food Agric., 2016, 96, 2952–2962 CrossRef CAS.
- A. Kim, S.-Y. Lee and S.-K. Chung, Phytomedicine, 2022, 102, 154144 CrossRef CAS.
- S. Mirzaei, M. H. Gholami, A. Zabolian, H. Saleki, M. V. Farahani, S. Hamzehlou, F. B. Far, S. O. Sharifzadeh, S. Samarghandian and H. Khan, Pharmacol. Res., 2021, 171, 105759 CrossRef CAS.
- A. Koraneekit, T. Limpaiboon, A. Sangka, P. Boonsiri, S. Daduang and J. Daduang, Oncol. Lett., 2018, 15, 7397–7402 Search PubMed.
- S.-R. Park, S.-R. Kim, I.-S. Hong and H.-Y. Lee, Front. Cell Dev. Biol., 2020, 8, 585987 CrossRef PubMed.
- C. Yuan, M.-H. Wang, F. Wang, P.-Y. Chen, X.-G. Ke, B. Yu, Y.-F. Yang, P.-T. You and H.-Z. Wu, Life Sci., 2021, 270, 119105 CrossRef CAS.
- Y. Sawata, T. Matsukawa, S. Doi, T. Tsunoda, N. Arikawa, N. Matsunaga, K. Ohnuki, S. Shirasawa and Y. Kotake, Mol. Cell. Biochem., 2019, 462, 25–31 CrossRef CAS.
- J. M. Brimson, M. I. Prasanth, D. S. Malar, P. Thitilertdecha, A. Kabra, T. Tencomnao and A. Prasansuklab, Pharmaceuticals, 2021, 14, 982 CrossRef CAS PubMed.
- A. Sgarbossa, D. Giacomazza and M. Di Carlo, Nutrients, 2015, 7, 5764–5782 CrossRef CAS PubMed.
- Y. P. Singh, H. Rai, G. Singh, G. K. Singh, S. Mishra, S. Kumar, S. Srikrishna and G. Modi, Eur. J. Med. Chem., 2021, 215, 113278 CrossRef CAS.
- Y. Dong, T. Stewart, L. Bai, X. Li, T. Xu, J. Iliff, M. Shi, D. Zheng, L. Yuan and T. Wei, Theranostics, 2020, 10, 179 CrossRef CAS.
- S. Huang, W. Liu, Y. Li, K. Zhang, X. Zheng, H. Wu and G. Tang, ACS Chem. Neurosci., 2021, 12, 419–429 CrossRef CAS.
- X. Dong and D. Zhao, CNS Neurosci. Ther., 2023, 29, 2397–2412 CrossRef CAS PubMed.
- A. Bumrungpert, S. Lilitchan, S. Tuntipopipat, N. Tirawanchai and S. Komindr, Nutrients, 2018, 10, 713 CrossRef.
- D. Li, Y. X. Rui, S. D. Guo, F. Luan, R. Liu and N. Zeng, Life Sci., 2021, 284, 119921 CrossRef CAS.
- Y. Wu, M. H. Wang, T. Yang, T. T. Qin, L. L. Qin, Y. M. Hu, C. C. Zhang, B. B. Sun, L. Ding and L. L. Wu, Front. Nutr., 2022, 8, 794841 CrossRef.
- H. Silva and N. M. F. Lopes, Front. Physiol., 2020, 11, 595516 CrossRef.
- B. Ramesh, P. Viswanathan and K. V. Pugalendi, Eur. J. Pharmacol., 2007, 566, 231–239 CrossRef CAS.
- O. Y. Althunibat, M. S. Abduh, M. H. Abukhalil, S. H. Aladaileh, H. Hanieh and A. M. Mahmoud, Biomed. Pharmacother., 2022, 149, 112900 CrossRef CAS.
- S. Shen, Y. Tong, Y. Luo, L. Huang and W. Gao, Nat. Prod. Rep., 2022, 39, 1856–1875 RSC.
- V. Křen and K. Valentová, Nat. Prod. Rep., 2022, 39, 1264–1281 RSC.
- Z. Bi, W. Zhang and X. Yan, Biomed. Pharmacother., 2022, 151, 113180 CrossRef CAS.
- Y. Fan, Q. Luo, J. Wei, R. Lin, L. Lin, Y. Li, Z. Chen, W. Lin and Q. Chen, Brain Res., 2018, 1679, 125–133 CrossRef CAS.
- T. Qin, A. Rasul, A. Sarfraz, I. Sarfraz, G. Hussain, H. Anwar, A. Riaz, S. Liu, W. Wei and J. Li, Int. J. Biol. Sci., 2019, 15, 2256–2264 CrossRef CAS PubMed.
- F. Annunziata, C. Pinna, S. Dallavalle, L. Tamborini and A. Pinto, Int. J. Mol. Sci., 2020, 21, 4618 CrossRef CAS PubMed.
- H. Deng, Q. Xu, H.-Y. Guo, X. Huang, F. Chen, L. Jin, Z.-S. Quan and Q.-K. Shen, Phytochemistry, 2022, 206, 113532 CrossRef.
- M. Jiang and H. Zhang, Curr. Opin. Biotechnol., 2016, 42, 1–6 CrossRef CAS.
- J. Zhang, A. ten Pierick, H. M. van Rossum, R. M. Seifar, C. Ras, J.-M. Daran, J. J. Heijnen and S. A. Wahl, Sci. Rep., 2015, 5, 12846 CrossRef CAS PubMed.
- Y. Deng and S. Lu, Crit. Rev. Plant Sci., 2017, 36, 257–290 CrossRef.
- K. M. Herrmann and L. M. Weaver, Annu. Rev. Plant Biol., 1999, 50, 473–503 CrossRef CAS PubMed.
- R. Patnaik and J. C. Liao, Appl. Environ. Microbiol., 1994, 60, 3903–3908 CrossRef CAS PubMed.
- M. Luttik, Z. Vuralhan, E. Suir, G. Braus, J. Pronk and J. Daran, Metab. Eng., 2008, 10, 141–153 CrossRef CAS PubMed.
- A. R. Hawkins, J. D. Moore and H. K. Lamb, Biochem. Soc. Trans., 1993, 21, 181–186 CrossRef CAS.
- K. Huang, M. Li, Y. Liu, M. Zhu, G. Zhao, Y. Zhou, L. Zhang, Y. Wu, X. Dai and T. Xia, Front. Plant Sci., 2019, 10, 1268 CrossRef.
- M.-H. Cho, O. R. Corea, H. Yang, D. L. Bedgar, D. D. Laskar, A. M. Anterola, F. A. Moog-Anterola, R. L. Hood, S. E. Kohalmi and M. A. Bernards, J. Biol. Chem., 2007, 282, 30827–30835 CrossRef CAS PubMed.
- H. Yoo, J. R. Widhalm, Y. Qian, H. Maeda, B. R. Cooper, A. S. Jannasch, I. Gonda, E. Lewinsohn, D. Rhodes and N. Dudareva, Nat. Commun., 2013, 4, 2833 CrossRef.
- A. Gloge, J. Zoń, Á. Kövári, L. Poppe and J. Rétey, Chem. –Eur. J., 2000, 6, 3386–3390 CrossRef CAS.
- C. B. Jendresen, S. G. Stahlhut, M. Li, P. Gaspar, S. Siedler, J. Förster, J. Maury, I. Borodina and A. T. Nielsen, Appl. Environ. Microbiol., 2015, 81, 4458–4476 CrossRef CAS PubMed.
- B. R. Albuquerque, S. A. Heleno, M. B. P. P. Oliveira, L. Barros and I. C. F. R. Ferreira, Food Funct., 2021, 12, 14–29 RSC.
- G. M. Walker and R. S. Walker, Adv. Appl. Microbiol., 2018, 105, 87–129 Search PubMed.
- N. Flores, J. Xiao, A. Berry, F. Bolivar and F. Valle, Nat. Biotechnol., 1996, 14, 620–623 CrossRef CAS.
- R. Patnaik and J. C. Liao, Appl. Environ. Microbiol., 1994, 60, 3903–3908 CrossRef CAS PubMed.
- S. Liu, J.-Z. Xu and W.-G. Zhang, World J. Microbiol. Biotechnol., 2022, 38, 22 CrossRef CAS.
- R. Chen, J. Gao, W. Yu, X. Chen, X. Zhai, Y. Chen, L. Zhang and Y. J. Zhou, Nat. Chem. Biol., 2022, 18, 520–529 CrossRef CAS.
- K. I. Minard and L. McAlister-Henn, J. Biol. Chem., 2005, 280, 39890–39896 CrossRef CAS PubMed.
- M. Deaner and H. S. Alper, Metab. Eng., 2017, 40, 14–22 CrossRef CAS.
- A. Bergman, V. Siewers, J. Nielsen and Y. Chen, AMB Express, 2016, 6, 115 CrossRef PubMed.
- G. M. Borja, A. Rodriguez, K. Campbell, I. Borodina, Y. Chen and J. Nielsen, Microb. Cell Fact., 2019, 18, 1–14 CrossRef CAS.
- P. P. Lin, A. J. Jaeger, T. Y. Wu, S. C. Xu, A. S. Lee, F. Gao, P. W. Chen and J. C. Liao, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 3538–3546 CrossRef CAS PubMed.
- G. M. Santangelo, Microbiol. Mol. Biol. Rev., 2006, 70, 253–282 CrossRef CAS PubMed.
- G. Gosset, J. Yong-Xiao and A. Berry, J. Ind. Microbiol., 1996, 17, 47–52 CrossRef CAS PubMed.
- Y. Lin and Y. Yan, Microb. Cell Fact., 2012, 11, 1–9 CrossRef PubMed.
- L. Guo, S. Ding, Y. Liu, C. Gao, G. Hu, W. Song, J. Liu, X. Chen and L. Liu, Biotechnol. Bioeng., 2022, 119, 983–993 CrossRef CAS.
- A. Rodriguez, K. R. Kildegaard, M. Li, I. Borodina and J. Nielsen, Metab. Eng., 2015, 31, 181–188 CrossRef CAS.
- D. Juminaga, E. E. Baidoo, A. M. Redding-Johanson, T. S. Batth, H. Burd, A. Mukhopadhyay, C. J. Petzold and J. D. Keasling, Appl. Environ. Microbiol., 2012, 78, 89–98 CrossRef CAS.
- F. Koopman, J. Beekwilder, B. Crimi, A. van Houwelingen, R. D. Hall, D. Bosch, A. J. van Maris, J. T. Pronk and J.-M. Daran, Microb. Cell Fact., 2012, 11, 1–15 CrossRef PubMed.
- M. Cai, Y. Wu, H. Qi, J. He, Z. Wu, H. Xu and M. Qiao, ACS Synth. Biol., 2021, 10, 49–62 CrossRef CAS.
- M. Reifenrath and E. Boles, Metab. Eng., 2018, 45, 246–254 CrossRef CAS.
- M. Li, K. R. Kildegaard, Y. Chen, A. Rodriguez, I. Borodina and J. Nielsen, Metab. Eng., 2015, 32, 1–11 CrossRef.
- M. Tatarko and T. Romeo, Curr. Microbiol., 2001, 43, 26–32 CrossRef CAS.
- J. Pittard, H. Camakaris and J. Yang, Mol. Microbiol., 2005, 55, 16–26 CrossRef CAS PubMed.
- T. Lütke-Eversloh and G. Stephanopoulos, Appl. Microbiol. Biotechnol., 2007, 75, 103–110 CrossRef.
- B. Kim, R. Binkley, H. U. Kim and S. Y. Lee, Biotechnol. Bioeng., 2018, 115, 2554–2564 CrossRef CAS PubMed.
- D. Na, S. M. Yoo, H. Chung, H. Park, J. H. Park and S. Y. Lee, Nat. Biotechnol., 2013, 31, 170–174 CrossRef CAS.
- K. Nijkamp, N. van Luijk, J. A. de Bont and J. Wery, Appl. Microbiol. Biotechnol., 2005, 69, 170–177 CrossRef CAS.
- J. Abatemarco, M. F. Sarhan, J. M. Wagner, J.-L. Lin, L. Liu, W. Hassouneh, S.-F. Yuan, H. S. Alper and A. R. Abate, Nat. Commun., 2017, 8, 332 CrossRef.
- Y. Liu, Y. Zhuang, D. Ding, Y. Xu, J. Sun and D. Zhang, ACS Synth. Biol., 2017, 6, 837–848 CrossRef CAS.
- M. Larroude, J. M. Nicaud and T. Rossignol, Microb. Biotechnol., 2021, 14, 2420–2434 CrossRef CAS.
- E. Kurpejović, A. Burgardt, G. M. Bastem, N. Junker, V. F. Wendisch and B. S. Akbulut, J. Biotechnol., 2023, 363, 8–16 CrossRef.
- I. Limem, E. Guedon, A. Hehn, F. Bourgaud, L. C. Ghedira, J.-M. Engasser and M. Ghoul, Process Biochem., 2008, 43, 463–479 CrossRef CAS.
- F. Zhang, J. Ren and J. Zhan, Appl. Biochem. Biotechnol., 2021, 193, 1099–1115 CrossRef CAS PubMed.
- H. B. Bang, K. Lee, Y. J. Lee and K. J. Jeong, Process Biochem., 2018, 68, 30–36 CrossRef CAS.
- J. Son, J. H. Jang, I. H. Choi, C. G. Lim, E. J. Jeon, H. Bae Bang and K. J. Jeong, Microb. Cell Factories, 2021, 20, 1–12 CrossRef.
- S. Masuo, Y. Kobayashi, K.-I. Oinuma and N. Takaya, Appl. Microbiol. Biotechnol., 2016, 100, 8701–8709 CrossRef CAS.
- K. Kukil and P. Lindberg, Microb. Cell Fact., 2022, 21, 1–16 CrossRef.
- Z. Qian, J. Yu, X. Chen, Y. Kang, Y. Ren, Q. Liu, J. Lu, Q. Zhao and M. Cai, ACS Synth. Biol., 2022, 11, 1600–1612 CrossRef CAS.
- N. Schultheiss, M. Roe and S. X. Boerrigter, CrystEngComm, 2011, 13, 611–619 RSC.
- L. Achnine, E. B. Blancaflor, S. Rasmussen and R. A. Dixon, Plant Cell, 2004, 16, 3098–3109 CrossRef CAS.
- T. Vannelli, W. W. Qi, J. Sweigard, A. A. Gatenby and F. S. Sariaslani, Metab. Eng., 2007, 9, 142–151 CrossRef CAS.
- K. Nijkamp, R. M. Westerhof, H. Ballerstedt, J. A. De Bont and J. Wery, Appl. Microbiol. Biotechnol., 2007, 74, 617–624 CrossRef CAS.
- Y. Kawai, S. Noda, C. Ogino, Y. Takeshima, N. Okai, T. Tanaka and A. Kondo, Microb. Cell Fact., 2013, 12, 1–9 CrossRef.
- Y. Huo, F. Wu, G. Song, R. Tu, W. Chen, E. Hua and Q. Wang, Chin. J. Biotech., 2020, 36, 2367–2376 CAS.
- Y. Brack, C. Sun, D. Yi and U. T. Bornscheuer, ChemBioChem, 2022, 23, e202200062 CrossRef CAS.
- Y. Huang, X. Jiang, W. Chen, G. Zhang and Q. Wang, Chin. J. Biotech., 2022, 38, 4553–4566 CAS.
- S. Zhang, J. Zhou, G. Zhang and J. Chen, Chin. J. Biotech., 2020, 36, 1838–1848 CAS.
- Y. Li, J. Li, B. Qian, L. Cheng, S. Xu and R. Wang, Molecules, 2018, 23, 3185 CrossRef.
- Q. Liu, T. Yu, X. Li, Y. Chen, K. Campbell, J. Nielsen and Y. Chen, Nat. Commun., 2019, 10, 4976 CrossRef CAS PubMed.
- L. F. Brey, A. J. Włodarczyk, J. F. B. Thøfner, M. Burow, C. Crocoll, I. Nielsen, A. J. Z. Nielsen and P. E. Jensen, Metab. Eng., 2020, 57, 129–139 CrossRef CAS.
- E.-B. Gao, K. Kyere-Yeboah, J. Wu and H. Qiu, Algal Res., 2021, 54, 102180 CrossRef.
- H. Qi, Y. Li, M. Cai, J. He, J. Liu, X. Song, Z. Ma, H. Xu and M. Qiao, J. Appl. Microbiol., 2022, 133, 707–719 CrossRef CAS.
- A. Rodriguez, Y. Chen, S. Khoomrung, E. Özdemir, I. Borodina and J. Nielsen, Metab. Eng., 2017, 44, 265–272 CrossRef CAS PubMed.
- R. Pereira, E. T. Mohamed, M. S. Radi, M. J. Herrgård, A. M. Feist, J. Nielsen and Y. Chen, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 27954–27961 CrossRef CAS.
- J. Combes, E. C. Rivera, T. Clément, C. Fojcik, V. Athes, M. Moussa and F. Allais, Sep. Purif. Technol., 2021, 259, 118170 CrossRef CAS.
- P. Zhang, W. Wenping, Z. Ying and Y. Bangce, Synth. Biol. J., 2021, 2, 778 Search PubMed.
- Y. Li, J. Mao, Q. Liu, X. Song, Y. Wu, M. Cai, H. Xu and M. Qiao, ACS Synth. Biol., 2020, 9, 756–765 CrossRef CAS PubMed.
- M. Berner, D. Krug, C. Bihlmaier, A. Vente, R. Müller and A. Bechthold, J. Bacteriol., 2006, 188, 2666–2673 CrossRef CAS.
- T. Furuya, Y. Arai and K. Kino, Appl. Environ. Microbiol., 2012, 78, 6087–6094 CrossRef CAS PubMed.
- Z. Chen, X. Sun, Y. Li, Y. Yan and Q. Yuan, Metab. Eng., 2017, 39, 102–109 CrossRef CAS PubMed.
- K. Sakae, D. Nonaka, M. Kishida, Y. Hirata, R. Fujiwara, A. Kondo, S. Noda and T. Tanaka, Enzyme Microb. Technol., 2023, 110193 CrossRef CAS PubMed.
- T. M. Louie, X. S. Xie and L. Xun, Biochemistry, 2003, 42, 7509–7517 CrossRef CAS PubMed.
- L. Liu, H. Liu, W. Zhang, M. Yao, B. Li, D. Liu and Y. Yuan, Engineering, 2019, 5, 287–295 CrossRef CAS.
- L. Wang, N. Li, S. Yu and J. Zhou, Bioresour. Technol., 2023, 368, 128320 CrossRef CAS.
- H. Qi, L. Yu, Y. Li, M. Cai, J. He, J. Liu, L. Hao, H. Xu and M. Qiao, Front. Microbiol., 2022, 13, 851706 CrossRef.
- T. Furuya and K. Kino, Appl. Microbiol. Biotechnol., 2014, 98, 1145–1154 CrossRef CAS PubMed.
- H. Kawaguchi, Y. Katsuyama, D. Danyao, P. Kahar, S. Nakamura-Tsuruta, H. Teramura, K. Wakai, K. Yoshihara, H. Minami and C. Ogino, Appl. Microbiol. Biotechnol., 2017, 101, 5279–5290 CrossRef CAS PubMed.
- X.-H. Wang, C. Zhao, X.-Y. Lu, H. Zong and B. Zhuge, ACS Synth. Biol., 2022, 11, 900–908 CrossRef CAS.
- Z. Zhuang, F. Song, H. Zhao, L. Li, J. Cao, E. Eisenstein, O. Herzberg and D. Dunaway-Mariano, Biochemistry, 2008, 47, 2789–2796 CrossRef CAS.
- H. Zhang and G. Stephanopoulos, Appl. Microbiol. Biotechnol., 2013, 97, 3333–3341 CrossRef CAS.
- D. M. De Oliveira, A. Finger-Teixeira, T. Rodrigues Mota, V. H. Salvador, F. C. Moreira-Vilar, H. B. Correa Molinari, R. A. Craig Mitchell, R. Marchiosi, O. Ferrarese-Filho and W. Dantas dos Santos, Plant Biotechnol. J., 2015, 13, 1224–1232 CrossRef CAS PubMed.
- A. Pandi and V. M. Kalappan, Mol. Biol. Rep., 2021, 48, 3733–3745 CrossRef CAS PubMed.
- O. Choi, C.-Z. Wu, S. Y. Kang, J. S. Ahn, T.-B. Uhm and Y.-S. Hong, J. Ind. Microbiol. Biotechnol., 2011, 38, 1657–1665 CrossRef CAS PubMed.
- K. Sun-Young, C. Oksik, L. Jae, H. Bang, U. Tai-Boong and H. Young-Soo, Microb. Cell Fact., 2012, 11, 153 CrossRef PubMed.
- J. L. Rodrigues, D. Gomes and L. R. Rodrigues, Front. Bioeng. Biotechnol., 2020, 8, 59 CrossRef PubMed.
- H. Lv, Y. Zhang, J. Shao, H. Liu and Y. Wang, Bioresour. Bioprocess., 2021, 8, 1–12 CrossRef.
- Z. Zhou, X. Zhang, J. Wu, X. Li, W. Li, X. Sun, J. Wang, Y. Yan, X. Shen and Q. Yuan, Metab. Eng., 2022, 73, 247–255 CrossRef CAS PubMed.
- M. Tylichová, D. Kopečný, S. Moréra, P. Briozzo, R. Lenobel, J. Snégaroff and M. Šebela, J. Mol. Biol., 2010, 396, 870–882 CrossRef PubMed.
- V. Vasiliou, A. Pappa and D. R. Petersen, Chem. Biol. Interact., 2000, 129, 1–19 CrossRef CAS PubMed.
- F. Lambert, J. Zucca, F. Ness and M. Aigle, Flavour Fragrance J., 2014, 29, 14–21 CrossRef CAS.
- S. Liu, L. Soomro, X. Wei, X. Yuan, T. Gu, Z. Li, Y. Wang, Y. Bao, F. Wang and B. Wen, Bioresour. Technol., 2021, 332, 124967 CrossRef CAS PubMed.
- J. Zhang, S. Liu, H. Sun, Z. Jiang, Z. Zhou, X. Han, Y. Zhou, H. Sun, W. Zhou and J. Mao, Foods, 2021, 10, 2577 CrossRef CAS PubMed.
- H. K. Sibhatu, S. A. Jabasingh, A. Yimam and S. Ahmed, LWT--Food Sci. Technol., 2021, 135, 110009 CrossRef CAS.
- H. B. Bang, Y. H. Lee, S. C. Kim, C. K. Sung and K. J. Jeong, Microb. Cell Fact., 2016, 15, 1–12 CrossRef PubMed.
- S. Liu, Q. Qi, N. Chao, J. Hou, G. Rao, J. Xie, H. Lu, X. Jiang and Y. Gai, Microb. Cell Fact., 2015, 14, 1–14 CrossRef CAS PubMed.
- M. Gottardi, J. D. Knudsen, L. Prado, M. Oreb, P. Branduardi and E. Boles, Appl. Microbiol. Biotechnol., 2017, 101, 4883–4893 CrossRef CAS PubMed.
- L. N. Jayakody and Y. S. Jin, Appl. Microbiol. Biotechnol., 2021, 105, 2675–2692 CrossRef CAS PubMed.
- O. De Smidt, J. C. Du Preez and J. Albertyn, FEMS Yeast Res., 2008, 8, 967–978 CrossRef CAS PubMed.
- H. B. Bang, J. Son, S. C. Kim and K. J. Jeong, Metab. Eng., 2023, 76, 63–74 CrossRef CAS PubMed.
- K. Venkatesan and K. Srinivasan, Arkivoc, 2008, 16, 302–310 Search PubMed.
- C. Zhang, Y. Zang, P. Liu, Z. Zheng and J. Ouyang, J. Biotechnol., 2019, 302, 92–100 CrossRef CAS PubMed.
- E. Klumbys, Z. Zebec, N. J. Weise, N. J. Turner and N. S. Scrutton, Green Chem., 2018, 20, 658–663 RSC.
- C. Zhang, Q. Xu, H. Hou, J. Wu, Z. Zheng and J. Ouyang, Microb. Cell Factories, 2020, 19, 163 CrossRef CAS PubMed.
- S. Y. Park, D. Yang, S. H. Ha and S. Y. Lee, Biotechnol. Bioeng., 2022, 119, 946–962 CrossRef CAS PubMed.
- J. Yang, J. Liang, L. Shao, L. Liu, K. Gao, J.-L. Zhang, Z. Sun, W. Xu, P. Lin and R. Yu, Metab. Eng., 2020, 59, 44–52 CrossRef CAS PubMed.
- J.-L. Ferrer, M. Austin, C. Stewart Jr and J. Noel, Plant Physiol. Biochem., 2008, 46, 356–370 CrossRef CAS PubMed.
- J. P. Wang, B. Liu, Y. Sun, V. L. Chiang and R. R. Sederoff, Front. Plant Sci., 2019, 9, 1942 CrossRef PubMed.
- H. Zhou, S. Gao, W. Zeng and J. Zhou, Biochem. Eng. J., 2021, 168, 107953 CrossRef CAS.
- J. Aschenbrenner, P. Marx, J. Pietruszka and J. Marienhagen, ChemBioChem, 2019, 20, 949–954 CrossRef CAS.
- F. Bourgaud, A. Hehn, R. Larbat, S. Doerper, E. Gontier, S. Kellner and U. Matern, Phytochem. Rev., 2006, 5, 293–308 CrossRef CAS.
- B.-I. Shimizu, Front. Plant Sci., 2014, 5, 549 Search PubMed.
- R. Vanholme, L. Sundin, K. C. Seetso, H. Kim, X. Liu, J. Li, B. De Meester, L. Hoengenaert, G. Goeminne and K. Morreel, Nat. Plants, 2019, 5, 1066–1075 CrossRef CAS PubMed.
- G. Vialart, A. Hehn, A. Olry, K. Ito, C. Krieger, R. Larbat, C. Paris, B. i. Shimizu, Y. Sugimoto and M. Mizutani, Plant J., 2012, 70, 460–470 CrossRef CAS.
- K. Kai, M. Mizutani, N. Kawamura, R. Yamamoto, M. Tamai, H. Yamaguchi, K. Sakata and B. i. Shimizu, Plant J., 2008, 55, 989–999 CrossRef CAS PubMed.
- Y. Lin, X. Sun, Q. Yuan and Y. Yan, Metab. Eng., 2013, 18, 69–77 CrossRef CAS PubMed.
- S.-M. Yang, G. Y. Shim, B.-G. Kim and J.-H. Ahn, Microb. Cell Fact., 2015, 14, 1–12 CrossRef CAS PubMed.
- Y. Zhao, X. Jian, J. Wu, W. Huang, C. Huang, J. Luo and L. Kong, J. Biol. Eng., 2019, 13, 1–13 CrossRef CAS PubMed.
- Y. Lin, X. Shen, Q. Yuan and Y. Yan, Nat. Commun., 2013, 4, 2603 CrossRef PubMed.
- X. Shen, M. Mahajani, J. Wang, Y. Yang, Q. Yuan, Y. Yan and Y. Lin, Metab. Eng., 2017, 42, 59–65 CrossRef CAS PubMed.
- C. H. Zhao, R. K. Zhang, B. Qiao, B. Z. Li and Y. J. Yuan, Biochem. Eng. J., 2020, 160, 107634 CrossRef CAS.
- S. Gao, H. N. Yu, R. X. Xu, A. X. Cheng and H. X. Lou, Phytochemistry, 2015, 111, 48–58 CrossRef CAS.
- A. D. Shrivastava and D. B. Kell, Molecules, 2021, 26, 2065 CrossRef CAS PubMed.
- F. Li, L. Yuan, H. Lu, G. Li, Y. Chen, M. K. Engqvist, E. J. Kerkhoven and J. Nielsen, Nat. Catal., 2022, 5, 662–672 CrossRef.
- A. Madani, B. Krause, E. R. Greene, S. Subramanian, B. P. Mohr, J. M. Holton, J. L. Olmos Jr, C. Xiong, Z. Z. Sun, R. Socher, J. S. Fraser and N. Naik, Nat. Biotechnol., 2023, 41, 1099–1106 CrossRef CAS PubMed.
- Y. Wang, S. Liao, N. Guan, Y. Liu, K. Dong, W. Weber and H. Ye, Sci. Adv., 2020, 6, eabb9484 CrossRef CAS PubMed.
- M. Ikeda, Appl. Microbiol. Biotechnol., 2006, 69, 615–626 CrossRef CAS PubMed.
- T. Yu, Y. J. Zhou, M. Huang, Q. Liu, R. Pereira, F. David and J. Nielsen, Cell, 2018, 174, 1549–1558 CrossRef CAS PubMed.
- S. Kwak, J. H. Jo, E. J. Yun, Y. S. Jin and J. H. Seo, Biotechnol. Adv., 2019, 37, 271–283 CrossRef CAS PubMed.
- D. Liu, M. S. Sica, J. Mao, L. F. Chao and V. Siewers, ACS Synth. Biol., 2022, 11, 3228–3238 CrossRef CAS PubMed.
- P. Xu, L. Li, F. Zhang, G. Stephanopoulos and M. Koffas, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 11299–11304 CrossRef CAS PubMed.
- E. M. Zhao, Y. Zhang, J. Mehl, H. Park, M. A. Lalwani, J. E. Toettcher and J. L. Avalos, Nature, 2018, 555, 683–687 CrossRef CAS PubMed.
- E. S. Bensen, B. G. Yeung and G. S. Payne, Mol. Biol. Cell, 2001, 12, 13–26 CrossRef CAS PubMed.
- M. Suástegui, C. Yu Ng, A. Chowdhury, W. Sun, M. Cao, E. House, C. D. Maranas and Z. Shao, Metab. Eng., 2017, 42, 134–144 CrossRef PubMed.
- H. Vashistha, M. Kohram and H. Salman, Elife, 2021, 10, e64779 CrossRef CAS PubMed.
- Y. Taniguchi, P. J. Choi, G. W. Li, H. Chen, M. Babu, J. Hearn, A. Emili and X. S. Xie, Science, 2010, 329, 533–538 CrossRef CAS PubMed.
- Y. Xiao, C. H. Bowen, D. Liu and F. Zhang, Nat. Chem. Biol., 2016, 12, 339–344 CrossRef CAS PubMed.
- Z. Zhou, Y. Liu, Y. Feng, S. Klepin, L. S. Tsimring, L. Pillus, J. Hasty and N. Hao, Science, 2023, 380, 376–381 CrossRef CAS PubMed.
- L. Guo, M. Qi, C. Gao, C. Ye, G. Hu, W. Song, J. Wu, L. Liu and X. Chen, Metab. Eng., 2022, 73, 235–246 CrossRef CAS PubMed.
- L. Guo, W. Diao, C. Gao, G. Hu, Q. Ding, C. Ye, X. Chen, J. Liu and L. Liu, Nat. Catal., 2020, 3, 307–318 CrossRef CAS.
- C. Ge, Z. Yu, H. Sheng, X. Shen, X. Sun, Y. Zhang, Y. Yan, J. Wang and Q. Yuan, Nat. Commun., 2022, 13, 2182 CrossRef CAS PubMed.
- E. X. Wang, Y. Liu, Q. Ma, X. T. Dong, M. Z. Ding and Y. J. Yuan, Biotechnol. Lett., 2019, 41, 951–961 CrossRef CAS PubMed.
- C. V. Dinh and K. L. J. Prather, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 25562–25568 CrossRef CAS PubMed.
- K. B. Xavier and B. L. Bassler, Curr. Opin. Microbiol., 2003, 6, 191–197 CrossRef CAS PubMed.
- R. Kalluri and V. S. LeBleu, Science, 2020, 367, eaau6977 CrossRef CAS PubMed.
- N. Ozber, J. L. Watkins and P. J. Facchini, J. Ind. Microbiol. Biotechnol., 2020, 47, 815–828 CrossRef CAS PubMed.
- H. Maeda and N. Dudareva, Annu. Rev. Plant Biol., 2012, 63, 73–105 CrossRef CAS PubMed.
- K. Zhou, K. Qiao, S. Edgar and G. Stephanopoulos, Nat. Biotechnol., 2015, 33, 377–383 CrossRef CAS.
- Z. Li, X. Wang and H. Zhang, Metab. Eng., 2019, 54, 1–11 CrossRef CAS PubMed.
- S. M. Brooks, C. Marsan, K. B. Reed, S. F. Yuan, D. D. Nguyen, A. Trivedi, G. Altin-Yavuzarslan, N. Ballinger, A. Nelson and H. S. Alper, Nat. Commun., 2023, 14, 4448 CrossRef CAS PubMed.
- Y. Liu, X. Tu, Q. Xu, C. Bai, C. Kong, Q. Liu, J. Yu, Q. Peng, X. Zhou, Y. Zhang and M. Cai, Metab. Eng., 2018, 45, 189–199 CrossRef CAS PubMed.
- M. Urui, Y. Yamada, Y. Ikeda, A. Nakagawa, F. Sato, H. Minami and N. Shitan, Microb. Cell Fact., 2021, 20, 200 CrossRef CAS PubMed.
- R. Wang, S. Zhao, Z. Wang and M. A. Koffas, Curr. Opin. Biotechnol., 2020, 62, 65–71 CrossRef CAS PubMed.
- S. K. Aulakh, L. Sellés Vidal, E. J. South, H. Peng, S. J. Varma, L. Herrera-Dominguez, M. Ralser and R. Ledesma-Amaro, Nat. Chem. Biol., 2023, 19, 951–961 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3np00012e |
| This journal is © The Royal Society of Chemistry 2024 |