DOI:
10.1039/B109456D
(Paper)
CrystEngComm, 2001,
3, 262-264
Copper(II) clofibriates, Part II. A two-dimensional coordination polymer of Cu(clofibriate)2(3-pyridylmethanol)2†
Received
17th October 2001
, Accepted 14th November 2001
Abstract
A two-dimensional coordination polymer of [Cu(clof)2(3-pymeth)2]n1
(clof = clofibriate anion and 3-pymeth = 3-pyridylmethanol) has been synthesized and characterized by single crystal structural analysis. The coordination environment about the copper(II) atom is tetragonal bipyramidal. The compound crystallizes in the monoclinic system, space group P21/c with a = 15.206(3), b = 8.029(2), c = 14.338(3) Å, β = 113.64(3)°, V = 1603.6(6) Å3 and Z = 2.
Introduction
The coordination bonds between transition metal ions and nitrogen containing heterocyclic ligands have proved useful for the construction of solid-state architectures and inorganic crystal engineering.2 Phenoxyalkanoic acids play an important role in biological processes as commercial auxin herbicide and/or anti-inflammatory agents.3 Copper(II) phenoxyalkanoate complexes have been shown to have diverse stereochemistry.3,4 Clofibric acid {2-(4-chlorophenoxy)-2-methylpropionic acid or 2-(4-chlorophenoxy)isobutyric acid} is a very interesting anti-inflammatory agent. Four copper(II) clofibriate complexes were studied by X-ray diffraction methods. X-Ray diffraction analysis of [Cu2(clof)4(ampy)2] (ampy = 2-aminopyrimidine)5 and [Cu2(clof)4(MeOH)2]6 shows that the compounds are binuclear with square pyramidal geometry at each copper(II) centre. The two copper(II) atoms are bridged by four carboxylate groups in syn–syn configuration of four clofibriate anions, while the apical ligands are 2-aminopyrimidine in [Cu2(clof)4(ampy)2]5 and methanol molecules in [Cu2(clof)4(MeOH)2],6 respectively. The structures of [Cu(clof)2L2], where L = pyridine (py)6 or nicotinamide (nia),7 are mononuclear, and each
copper(II) atom has a tetragonal-bipyramidal environment with the CuO4N2 chromophore.
This paper present the preparation of [Cu(clof)2(3-pymeth)2]
1
[3-pymeth = ronicol (3-pyridylmethanol)] and its crystal and molecular structures.
Experimental
Preparation of the complex
1 was prepared by addition of 3-pyridylmethanol (0.02 mol) to copper(II) clofibriate (0.01 mol) in hot methanol.7 The mixture was stirred, filtered and left to cool and stand at room temperature. A blue product precipitated and was collected; recrystallization from methanol–acetone (4∶1) solution gave air stable blue crystals. Anal. Found: C, 57.5; H, 4.6; Cu, 9.0; N, 4.0. Calc. for [Cu(clof)2(3-pymeth)2]
1: C, 57.60; H, 4.55, Cu, 8.96, N, 3.95%.
Structure determination
Data collection at 100 K using an Oxford Cryosystems cooler device8 and cell refinement were carried out using a four-circle diffractometer Kuma KM4CCD with graphite monochromated MoKα radiation. The diffraction intensities were corrected for Lorentz and polarization factors. The structure was solved by heavy atom methods using SHELXS-97.9 Geometrical analysis was performed using SHELXL-9710 and PLATON-99.11 Empirical absorption correction was made by using XABS2.12 The structure of the complex was drawn by using ORTEP-3.13 The Single Crystal Suite WINGX was used as an integrated system for all the crystallographic programs.14 Crystal data and conditions of data collection and refinement are reported in Table 1.
|
Click b109456d.txt for full crystallographic data (CCDC 167248).
|
| Empirical formula |
C32H34Cl2N2O8Cu1 |
| Crystal dimensions/mm |
0.17 × 0.15 × 0.12 |
|
M
|
709.05 |
|
T/K |
100(2) |
| Crystal system |
Monoclinic |
| Space group |
P21/c |
|
a/Å |
15.206(3) |
|
b/Å |
8.029(2) |
|
c/Å |
14.338(3) |
|
β/° |
113.64(3) |
|
Z
|
2 |
| Absorption correction: Empirical |
T
min = 0.6945, Tmax = 1.0830 |
|
D
c/Mg m−3 |
1.467 |
|
μ(MoKα)/mm−1 |
0.901 |
|
F(000) |
734 |
|
θ Range/° |
3.58–27 |
| Reflections collected |
10025 |
| Independent reflections |
3443 [Rint = 0.0312] |
| Final R indices [I > 2σ(I)] |
R
1 = 0.0327, wR2 = 0.0787 |
|
R indices (all data) |
R
1 = 0.0424, wR2 = 0.0830 |
Results and discussion
The molecule of 1 lies about an inversion centre and the molecular structure of 1 is shown in Fig. 1. Selected bond lengths and bond angles are summarized in Table 2. The coordination environment of the copper(II) atom is tetragonal bipyramidal. The tetragonal plane is built up by a pair of unidentate clofibriate anions using carboxylate oxygen atoms [Cu–O1 = 1.9892(15) Å] and a pair of neutral ronicol molecules using nitrogen atoms of pyridine rings [Cu–N1 = 2.0077(15) Å] in trans positions. The axial positions are occupied by two hydroxyl oxygen atoms [Cu–O4iv = 2.4406(14) Å] from two adjacent molecules of
ronicol [x, −y − 1/2, z + 1/2]. The interatomic distance Cu⋯O2 is 3.282(2) Å, which should be regarded as non-bonding. In the mononuclear complexes [Cu(clof)2(py)2]6 and [Cu(clof)2(nia)2]7 with bidentate bi-coordinated carboxylic groups the bond lengths Cu–O2 are 2.683(1) Å and 2.614(2) Å, respectively. The intramolecular hydrogen bond O4iv–H17iv⋯O2 (Table 3) creates a six membered metalocycle and stabilizes the molecular structure. The torsion angle C1–C2–O3–C3 of −55.6(2)° is similar to the torsion angles in [Cu(clof)2(py)2]6 and [Cu(clof)2(nia)2]7 of −57.4° and −59.8°, respectively.
![Perspective view of complex [Cu(clof)2(3-pymeth)2]n1, with the atom numbering scheme. Thermal ellipsoids are drawn at the 50% probability level. Click image or here to access a 3-D representation.](/image/article/2001/CE/b109456d/b109456d-f1.gif) |
| | Fig. 1
Perspective view of complex [Cu(clof)2(3-pymeth)2]n1, with the atom numbering scheme. Thermal ellipsoids are drawn at the 50% probability level. Click image or 1.htm to access a 3-D representation.
| |
Table 2
Selected bond lengths (Å) and angles (°) for [Cu(clof)2(3-pymeth)2]
1
| Symmetry code: (i)
−x, −y, −z; (ii)
x, −y − 1/2, z + 1/2; (iii)
−x, y + 1/2, −z − 1/2; (iv)
−x, y − 1/2, −z − 1/2; (v)
x, −y + 1/2, +z + 1/2; (vi)
x, −y − 1/2, z − 1/2; (vii)
x, −y + 1/2, z − 1/2; (viii)
−x − 1, y − 1/2, −z − 1/2; (ix)
−x − 1, −y + 1, −z. |
| Cu–O1 |
1.9892(15) |
Cu–N1 |
2.0077(15) |
| Cu–O4iv |
2.4406(14) |
O1–C1 |
1.274(2) |
| O2–C1 |
1.243(2) |
O3–C3 |
1.362(2) |
| O3–C2 |
1.450(2) |
O4–C16 |
1.425(2) |
| C1–C2 |
1.549(3) |
|
|
| |
|
|
|
| O1i–Cu–O1 |
180.0 |
O1–Cu–N1 |
89.37(6) |
| O1–Cu–O4iv |
92.50(7) |
N1–Cu–O4iv |
94.05(6) |
| N1–Cu–N1i |
180.0 |
C1–O1–Cu |
127.4(1) |
| C3–O3–C2 |
121.1(1) |
C11–N1–C15 |
118.2(2) |
| C11–N1–Cu |
118.6(1) |
C15–N1–Cu |
123.2(1) |
| O2–C1–O1 |
126.7(2) |
O2–C1–C2 |
117.6(2) |
| O1–C1–C2 |
115.6(2) |
O3–C2–C1 |
110.6(1) |
| O4–C16–C12 |
109.5(2) |
Cu–O4iv–C16iv |
143.4(1) |
Table 3
Parameters of intermolecular interactions within the structure 1
| D–H⋯A |
d(D–H)/Å |
d(H⋯A)/Å |
d(D⋯A)/Å |
<(DHA)/° |
| Symmetry code: (iii)
−x, y + 1/2, −z − 1/2; (iv)
−x, y − 1/2, −z − 1/2; (vi)
x, −y − 1/2, z − 1/2; (vii)
x, −y + 1/2, z − 1/2; (viii)
−x − 1, y − 1/2, −z − 1/2; (ix)
−x − 1, −y + 1, −z. |
| O4iv–H17iv⋯O2 |
0.83(3) |
1.74(3) |
2.599(2) |
172(3) |
| C11–H11⋯O4iii |
0.95(2) |
2.52(2) |
3.033(2) |
113(2) |
| C16–H15⋯O2iii |
1.00(2) |
2.55(2) |
3.248(2) |
127(2) |
| C13–H12⋯O3iv |
0.93(2) |
2.66(2) |
3.462(2) |
147(2) |
| C8–H14⋯Clvi |
0.92(2) |
2.92(2) |
3.745(2) |
150(2) |
| C10–H8⋯Clviii |
0.95(2) |
3.01(2) |
3.847(3) |
147(2) |
| C6–Cl⋯Clix |
— |
3.359(1) |
— |
142.86(8) |
The crystal structure of 1 consists of 2-D sheets15
(Fig. 2). The layers are situated in the bc plane and consist of two zigzag chains crossing at the Cu(II) atom and several hydrogen-bonding interactions C–H⋯O,16
[C11–H11⋯O4iii; C16–H15⋯O2iii; C13–H12⋯O3iv]
(Table 3) as shown in Fig. 2. The weak hydrogen bonds are now intensively studied;16 the role of weak C–H⋯O hydrogen bonds in crystal engineering of organometallic complexes has been recently reviewed by Braga et al.17 In complex 1 C–H⋯O
hydrogen bonds have the role of stabilizing the 2-D chains. In Fig. 3 a view of the packing of 1 perpendicular to the ac plane is shown. As can be seen, the layers are connected by short Cl⋯Cl contacts18 and weak C–H⋯Cl interactions,19
[C8–H4⋯Clvi; C10–H8⋯Clviii]
(Table 3), resulting in a 3-D network.15 The intermolecular interactions C–H⋯Cl with organic chlorine acceptor groups were previously the subject of controversial discussion; Thallapally and Nangia19a suggested that C–H⋯Cl are essentially van der Waals contacts, on the other hand Brammer et al.19b suggested that
C–H⋯Cl are weak hydrogen bonds. In complex 1 the weak C–H⋯Cl interactions together with the short Cl⋯Cl interactions are dominant in the created 3-D network.
 |
| | Fig. 2
View perpendicular to the bc plane showing the hydrogen bonding; chlorine atoms and benzene rings of clofibriate anions are omitted for clarity.
| |
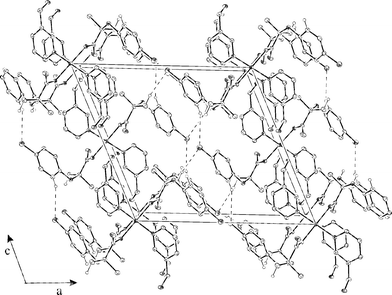 |
| | Fig. 3
View of the packing of 1 in the cell, viewed perpendicular to the ac plane; hydrogen atoms are omitted for clarity. Click image or 3.htm to access a 3-D representation.
| |
The crystal structure of the compound under study can be compared with those of [CuX2(3-pymeth)2]n, where X = salicylate (sal)20 or niflumate (nif; 2-[α,α,α-trifluoro-m-tolylamino]pyridine-3-carboxylate).21 The Cu–Oeq
(oxygen atom of carboxylic group), Cu–Neq
(nitrogen atom of pyridine ring of ronicol) and Cu–Oax
(oxygen atom of hydroxyl group of ronicol) lengths for the former structure are 1.944(4) Å, 2.039 Å and 2.622(4) Å, respectively. In the latter structure the values are 1.946(3) Å, 2.038(3) Å and 2.573(3) Å, respectively. All three compounds have polymeric structures with axially
elongated tetragonal bipyramidal geometry about each Cu(II) atom of the CuO4N2 chromophore, as predicted by the Jahn–Teller theorem. The T parameter (T = Rs/RL), indicating the degree of tetragonal distortion about the copper(II) centers,22 decreases in the sequence: 0.819 1 > 0.774 [Cu(nif)2(3-pymeth)2]n > 0.759 [Cu(sal)2(3-pymeth)2]n. Another structure variation for [CuX2(3-pymeth)2]n was found for [Cu(nif)2(3-pymeth)2]n.21
In this structure, ambidentate 3-pyridylmethanol molecules in the 1-D infinite chains bridge copper(II) atoms.21
Conclusions
Analysis of the packing of molecules of 1 in the crystal shows that the strong O–H⋯O as well as the weak C–H⋯O interactions play a role in the stabilization of 2-D coordination polymers. The interlayer contacts are found to dominate in the absence of the strong hydrogen bonds. Short Cl⋯Cl and weak C–H⋯Cl interactions have been observed, which link layers of the 2-D coordination polymer.
Acknowledgements
This work was financially supported by grant 1/6106/20 from the Slovak Grant Agency for Science.
References
- M. Melník, M. Koman, D. Hudecová, J. Moncol', B. Dudová, T. Glowiak, J. Mroziński and C. E. Holloway, Inorg. Chim. Acta, 2000, 308, 1 CrossRef CAS.
- See, for example: D. Braga, J. Chem. Soc., Dalton Trans., 2000, 3705 Search PubMed; A. J. Blake, N. R. Champness, P. Hubberstey, W.-S. Li, A. Withersby and M. Schröder, Coord. Chem. Rev., 1999, 183, 117 RSC; G. F. Swiegers and T. J. Malefetse, Chem. Rev., 2001, 100, 3483 CrossRef CAS; J. A. R. Navarro and B. Lippert, Coord. Chem. Rev., 1999, 185–186, 653 CrossRef; A.
J. Blake, N. R. Champness, D. A. Leminovskii, A. G. Majouya, N. V. Zyk and M. Schröder, Coord. Chem. Rev., 2001, 222, 155 CrossRef CAS; J. A. R. Navarro and B. Lippert, Coord. Chem. Rev., 2001, 222, 219 CrossRef CAS.
- G. Smith, E. J. O'Reilly, C. H. L. Kennard and K. E. Brown, Inorg. Chim. Acta, 1981, 49, 53 CrossRef CAS; C. Dendrinou-Samara, G. Psomas, K. Christophorou, V. Tangoulis, C. P. Raptopoulou, A. Terzis and D. P. Kessissoglou, J. Chem. Soc., Dalton Trans., 1996, 3737 RSC; G. Smith, E. J. O'Reilly, C. H. L. Kennard and A. H. White, J. Chem. Soc., Dalton Trans., 1985, 243 RSC; G. Smith, E. J. O'Reilly, C. H. L. Kennard, K. Stadnicka and B. Oleskyn, Inorg. Chim. Acta, 1981, 47, 111 CrossRef CAS.
- G. Smith, D. Lynch, T. C. W. Mak and W.-H. Yip, Polyhedron, 1993, 12, 203 CrossRef CAS; G. Smith, E. J. O'Reilly, C. H. L. Kennard and T. C. W. Mak, Inorg. Chim. Acta, 1982, 65, L219 CrossRef CAS; G. Smith, E. J. O'Reilly and C. H. L. Kennard, Inorg. Chim. Acta, 1981, 62, 241 CrossRef; C. H. L. Kennard, S. W. Steward, E. J. O'Reilly, G. Smith and A. H. White, Polyhedron, 1985, 4, 697 CrossRef CAS; E. J. O'Reilly, G. Smith and C. H. L. Kennard, Inorg. Chim. Acta, 1984, 90, 63 CrossRef CAS; T. C. W. Mak, W.-H. Yip, G. Smith, E. J. O'Reilly and C. H. L. Kennard, Inorg. Chim. Acta, 1986, 112, 53 CrossRef CAS; G. Psomas, C. P. Raptopoulou, L. Iordanidis, C. Dendrinou-Samara, C. Tangoulis and D. P. Kessissoglou, Inorg. Chem., 2000, 39, 3042 CrossRef CAS; C. H. L. Kennard, E. J. O'Reilly, S. Schiller, G. Smith and A. H. White, Aust. J. Chem., 1986, 39, 1823 CAS; R. Stomberg and K. Lundquist, Acta Chem. Scand., 1989, 43, 160 CAS; G. Psomas, C. Dendrinou-Samara, P. Philippakopoulos, V. Tangoulis, C. P. Raptopoulou, E. Samaras and D. P. Kessissoglou, Inorg. Chim. Acta, 1998, 272, 24 CrossRef CAS; C. Dendrinou-Samara, G. Psomas, C. P. Raptopoulou and D. P. Kessissoglou, J. Inorg. Biochem., 2001, 83, 7 CrossRef CAS; G. Smith, E. J. O'Reilly, H. L. Carrell, C. J. Carrell and C. H. L. Kennard, Polyhedron, 1996, 15, 1995 CrossRef CAS; G. Smith and C. H. L. Kennard, Inorg. Chim. Acta, 1989, 161, 3 CrossRef CAS.
- T. C. W. Mak, C. H. L. Kennard, G. Smith, E. J. O’Reilly, D. S. Sagatys and J. C. Fulwood, Polyhedron, 1987, 6, 855 CrossRef CAS.
- Y. Kani, S. Ohba, H. Matsushima and T. Tokii, Acta Crystallogr., Sect. C, 1998, 54, 193 CrossRef.
- M. Melník, M. Koman, D. Hudecová, J. Moncol', B. Dudová, T. Glowiak, J. Mroziński and C. E. Holloway, Inorg. Chim. Acta, 2000, 308, 1 CrossRef CAS.
- J. Cosier and A. M. Glazer, J. Appl. Crystallogr., 1986, 19, 105 CrossRef CAS.
- G. M. Sheldrick, Acta Crystallogr., Sect. A, 1990, 46, 467 CrossRef.
-
G. M. Sheldrick, SHELXL-97, Program for the Refinement of Crystal Structures, University of Goettingen, Germany, 1997.
-
A. L. Spek, PLATON-99, Molecular Geometry Program, University of Utrecht, The Netherlands, 1999.
- S. Parkin, B. Moezzi and H. Hope, J. Appl. Crystallogr., 1995, 28, 53 CrossRef CAS.
- L. J. Farrugia, J.
Appl. Crystallogr., 1997, 30, 565 Search PubMed.
- L. J. Farrugia, J. Appl. Crystallogr., 1999, 32, 837 CrossRef.
- K. S. Min and M. P. Suh, Eur. J. Inorg. Chem., 2001, 449 CrossRef CAS.
-
G. R. Desiraju and T. Steiner, The Weak Hydrogen Bond, Oxford University Press, Oxford, UK, 1999 Search PubMed; W. Yang, C. Lu, C. Wu, D. Wu and H. Zhuang, Inorg. Chem. Commun., 2001, 4, 285 Search PubMed; B. Ahrens, S. Friedrichs, R. Herbst-Irmer and P. G. Jones, Eur. J. Inorg. Chem., 2000, 2017 CrossRef CAS.
- D. Braga, F. Grepioni and G. R. Desiraju, Chem. Rev., 1998, 98, 1375 CrossRef CAS.
- G. R. Desiraju, Angew. Chem., Int. Ed. Engl., 1995, 34, 3211 CrossRef; I. Csoregh, T. Brehmer, P. Bombicz and E. Weber, Cryst. Eng., 2001, 4, 343 CrossRef CAS.
-
(a) P. K. Thallapally and A. Nangia, CrystEngComm, 2001, 27, RSC;
(b) L. Brammer, E. A. Bruton and P. Sherwood, Cryst. Growth Des., 2001, 1, 277 Search PubMed.
- N. N. Hoang, F. Valach, L'. Macášková and M. Melník, Acta Crystallogr., Sect. C, 1992, 48, 1933 CrossRef.
- F. Valach, M. Tokarčik, P. Kubinec, M. Melník and L'. Macášková, Polyhedron, 1997, 16, 1461 CrossRef CAS.
- J. Gažo, I. B. Bersuker, J. Garaj, M. Kabešová, J. Kohout, H. Langfelderová, M. Melník, M. Serátor and F. Valach, Coord. Chem. Rev., 1976, 19, 253 CrossRef CAS.
|
| This journal is © The Royal Society of Chemistry 2001 |
Click here to see how this site uses Cookies. View our privacy policy here. ![Perspective view of complex [Cu(clof)2(3-pymeth)2]n1, with the atom numbering scheme. Thermal ellipsoids are drawn at the 50% probability level. Click image or here to access a 3-D representation.](/image/article/2001/CE/b109456d/b109456d-f1.gif)


