A novel host framework of cholic acid inclusion crystals by slide and flip of the layers
Nungruethai
Yoswathananont
*a,
Suwabun
Chirachanchai
b,
Kohji
Tashiro
c,
Kazunori
Nakano
d,
Kazuki
Sada
a and
Mikiji
Miyata
a
aMaterial and Life Science, Graduate School of Engineering, Yamadaoka, Suita, Osaka University, Osaka, 565-0871, Japan. E-mail: nun@ap.chem.eng.osaka-u.ac.jp; miyata@ap.chem.eng.osaka-u.ac.jp
bThe Petroleum and Petrochemical College, Chulalongkorn University, Bangkok, 10330, Thailand
cGraduate School of Science, Osaka University, Toyonaka, Osaka, 560-0043, Japan
dNagoya Municipal Industrial Research Institute, Rokuban, Atsuta-ku, Nagoya, 456-0058, Japan
Abstract
A novel host framework of cholic acid (CA) has been observed in the crystal structure of m-chloroaniline clathrate. Crystallographic study reveals that CA forms a bilayer-type structure; however, slide and flip of the lipophilic layers give a different host framework to those of over 100 inclusion crystals previously reported. Structural comparison between the clathrates of aniline and m-chloroaniline indicates that the addition of a chlorine atom leads to isomerization of the open host framework due to a steric effect between adjacent guests within the host cavities.
Introduction
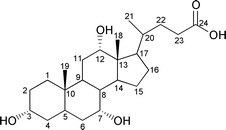
Cholic acid (CA), one of the steroidal bile acids, is known to form inclusion compounds with a wide variety of organic substances.1–5 Its molecular structure and numbering scheme are shown below.The most striking of these inclusion compounds has a facially amphiphilic structure which is due to three hydroxy groups in the steroidal nucleus pointing α to (i.e. below) the plane of the ring, forming a hydrophilic face, and two methyl groups which are β-orientated (pointing above the plane of the ring) forming a lipophilic face. X-Ray crystallographic studies have revealed that CA forms bilayer structures via an alternating stack of hydrophilic and lipophilic layers and that host cavities are formed in the lipophilic layers due to the bent molecular shape of the component CA molecules. It should be noted that there are many possibilities for the stacking of these layers but for simplicity we consider two variations on the lipophilic side. One arises through flip of the layers, parallel and antiparallel, as shown in Fig. 1. The other arises through sliding (α-, β- and non interdigitation-type) between the layers (Fig. 2), where the type of sliding is characterized by the interdigitation patterns of the methyl groups within the lipophilic layers. The combination of flip and sliding between the layers can produce at least six types of stacking arrangement, as shown in Table 1; three of these have already been found.4e,6,7 Here, we present the novel crystal structure of CA with m-chloroaniline which shows a unique parallel orientation with non interdigitation-type sliding in the lipophilic layer.
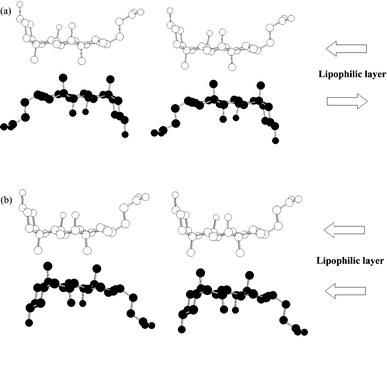 | ||
| Fig. 1 Schematic representation of the molecular orientation (a) antiparallel and (b) parallel in the lipophilic layer. The two ends of the molecule, that of the hydroxy group at the carbon 3-position and that of the carboxylic group, are distinguished as the head and tail, respectively. The heads of the arrows represent the head of cholic acid. | ||
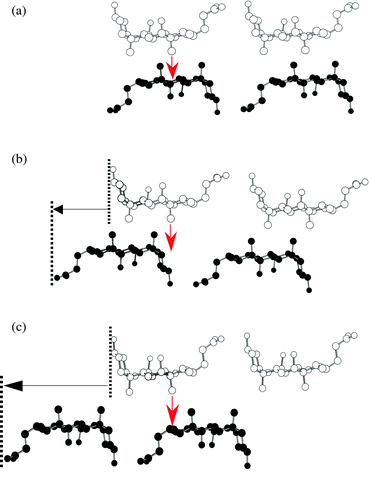 | ||
| Fig. 2 Schematic representation of sliding types for the lipophilic layers: (a) α-, (b) β- and (c) non interdigitation-type. | ||
Results and discussion
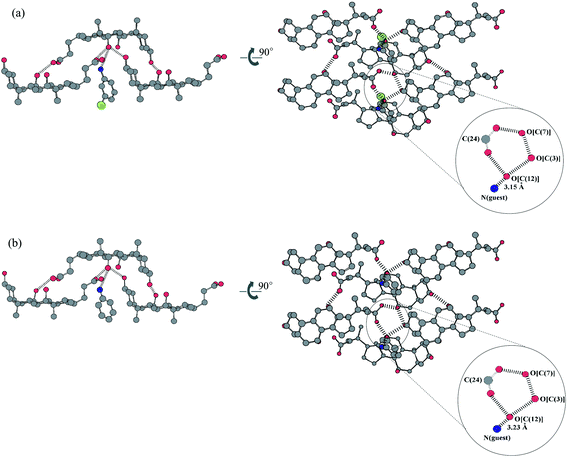
Platelet-like crystals were obtained by recrystallizing CA and m-chloroaniline from a butan-1-ol solution. The 1∶1 host∶guest stoichiometry was confirmed by thermal analysis. X-Ray crystallographic studies revealed that the crystal belongs to the orthorhombic P212121 space group; their crystal data are given in Table 2. The crystal packing diagram is shown in Fig. 3(a), together with that of CA with aniline4a,7 for comparison in Fig. 3(b). The crystal structures of the m-chloroaniline clathrate and aniline clathrate are different, although in the hydrophilic layer both compounds have the same stacking pattern and form the same cyclic hydrogen bond networks as shown in Fig. 4. In this layer, amino groups in both clathrates are found to form weak hydrogen bonds with the hydroxy groups of the host at distances of 3.15 and 3.23 Å, respectively.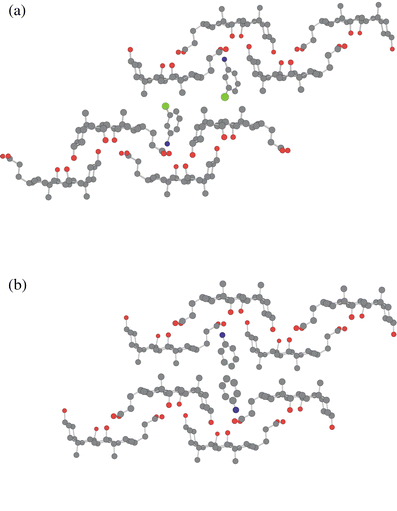 | ||
| Fig. 3 Molecular packing diagrams of CA with (a) m-chloroaniline and (b) aniline, viewed along the crystallographic c- and b-axes, respectively. Carbon, oxygen, nitrogen and chlorine are represented in grey, red, blue and green, respectively. Hydrogen atoms are omitted for clarity. Click image or 3a.htm to access a 3D representation for (a). Click image or 3b.htm to access a 3D representation for (b). | ||
 | ||
| Fig. 4 Hydrogen bond networks and the orientation of guest molecules within the channel of the hydrophilic layer for (a) m-chloroaniline and (b) aniline. Carbon, oxygen, nitrogen and chlorine are represented in grey, red, blue and green, respectively. Hydrogen atoms are omitted for clarity. | ||
| Parameter | CA∶m-chloroaniline |
|---|---|
| a Click b101906f.txt for full crystallographic data (CCDC 161956). | |
| Host∶guest | 1∶1 |
| Empirical formula | C30H46O5NCl |
| M | 536.15 |
| Crystal system | Orthorhombic |
| Space group | P212121 |
| a/Å | 14.6238(8) |
| b/Å | 24.903(2) |
| c/Å | 8.0556(3) |
| V/Å3 | 2933.6(3) |
| Z | 4 |
| T/K | 203.2 |
| D c/g cm−3 | 1.21 |
| μ/mm−1 | 1.45 |
| Number of unique reflections | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 081 081 |
| Number of observed reflections | 2997 |
| R 1 | 0.064 |
On the other hand, in the lipophilic layer, the m-chloroaniline clathrate shows a different stacking mode to that of the aniline clathrate and other bilayer structures. There are no interdigitations of the two methyl groups (C18 and C19) between the upper and the lower layers, and the chlorine atom of the guest component interdigitates with the two methyl groups in the lower layers, Fig. 3(a). Comparing this with the aniline clathrate, Fig. 3(b), the methyl groups (C19) in the upper layer are interdigitated between the methyl groups (C18 and C19) of the opposite layer; this type of interdigitation is classified as α-type. (Note that the meaning of α with regard to interdigitation type is different from that used earlier when describing functional group orientation within CA.) The interdigitation here of the methyl groups in the lipophilic layer gives rise to the formation of a one-dimensional host channel. Moreover, orientations of the layers in both clathrates are different. The m-chloroaniline clathrate gives parallel orientation of the CA molecules in the lipophilic layer and antiparallel in the hydrophilic layer, whereas the aniline clathrate gives antiparallel orientation of CA molecules in both layers. The flip of the layer and the stacking pattern of the m-chloroaniline clathrate differ from all previously reported crystal structures of CA inclusion compounds.
It should be noted that in both clathrates, positions and orientations of the guests in the host cavities are quite similar (Fig. 4). The same host–guest hydrogen bonds from the amino groups to the host compounds enable the orientations of the guest components to be fixed. Therefore, the addition of a chlorine atom into the aniline guest destabilizes the α-gauche type host framework of CA because of the repulsion induced by the steric effects introduced between the adjacent m-chloroaniline guest molecules. As a result, the host framework of CA is changed by slide and flip of the lipophilic layer.
This change in the lipophilic layer alters the host cavity from that of a channel-type (in aniline) to a two-cage-type (for m-chloroaniline). However, the size of the host cavity in the m-chloroaniline clathrate is larger than that of the aniline inclusion compound; Table 3 summarizes the volumes of the host cavities and their packing coefficients (PCcavity), together with the volume ratios of the guest components to host cavities. More recently, we have reported that PCcavity, the parameter that estimates the steric fit between the host and the guest, plays an important role in the isomerization of the bilayer structure of CA with a series of benzene derivatives.6 PCcavity values for both clathrates are similar and within the range for stable inclusion crystals (55–72%).6 In order to clarify the rationale for the change in host frameworks between the two, a value of PCcavity for the hypothetical inclusion crystal with m-chloroaniline in the α-gauche type host framework was calculated and found to be the same as that for the aniline clathrate. This value (67%) is also within the above range for stable inclusion compounds. From the viewpoint of the guest volume, it is possible to incorporate m-chloroaniline into the α-gauche type host framework (Table 3). However, all of the crystals that we obtained for m-chloroaniline clathrate exhibit the new type of host framework. This suggests that the shape and orientation of the guest compound induces isomerization of the host framework.
| Guest | Molecular volume of guest/Å3 | Number of guests per unit cell | Volume of host cavitya per unit cell/Å3 | Volume of host cavity per guest molecule/Å3 | PCcavityb (%) |
|---|---|---|---|---|---|
| a Volume of cavity per unit cell is calculated using a probe of radius 0.7 Å. b PCcavity is the packing coefficient of the guest component within the host cavity, given by the following expression: PCcavity (%)⊕=⊕[(Molecular volume of guest molecule)⊕×⊕(Number of guest molecules)⊕×⊕100]/Volume of host cavity. c Hypothetical guest for this framework type. | |||||
| Aniline (α-gauche type framework) | 95.8 | 2 | 336.0 | 168.0 | 57.0 |
| m-Chloroaniline | 112.0 | 4 | 786.2 | 196.5 | 57.0 |
| m-Chloroaniline (in α-gauche type frameworkc) | 112.0 | 2 | 336.0 | 168.0 | 66.6 |
In conclusion, we demonstrate a new, open host framework for the inclusion crystal structure of CA with m-chloroaniline which shows a novel stacking pattern in the lipophilic layer. The present work indicates that CA has flexible host frameworks to fit a wide range of organic compounds. Further systematic structural investigations of CA may give us other novel host frameworks.
Experimental
All chemicals and solvents were commercially available and used without further purification. Differential thermal analysis (DTA) and thermal gravimetry (TG) were performed using a Rigaku Thermo Plus TG-8120; ca. 10 mg of sample were used for each run over the temperature range 30–230![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) °C at a heating rate of 5
°C at a heating rate of 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) °C min−1. X-Ray powder diffraction (XRD) patterns were measured using a Rigaku RINT-1100 diffractometer at room temperature. XRDP: 7.02(40), 12.96(10), 14.12(100). Inclusion crystals were prepared by the same procedure as that described in a previous paper.6
°C min−1. X-Ray powder diffraction (XRD) patterns were measured using a Rigaku RINT-1100 diffractometer at room temperature. XRDP: 7.02(40), 12.96(10), 14.12(100). Inclusion crystals were prepared by the same procedure as that described in a previous paper.6
Crystal structure determination
X-Ray single crystal diffraction data were collected on a Rigaku R-AXIS RAPID diffractometer, using graphite-monochromatized Cu-Kα radiation. Lattice parameters were obtained by reflections from three oscillation images for the area detector. Direct methods (SIR-92)8 were employed for the structure solution. Absolute configurations conformed to the previously reported structure of CA.1a Structures were refined based on F![[hair space]](https://www.rsc.org/images/entities/char_200a.gif) 2 by a full-matrix, least squares procedure with the program teXsan.9 All non-hydrogen atoms were refined with anisotropic displacement parameters and hydrogen atoms were placed in idealized positions.
2 by a full-matrix, least squares procedure with the program teXsan.9 All non-hydrogen atoms were refined with anisotropic displacement parameters and hydrogen atoms were placed in idealized positions.
Calculations
The volume of the host cavity for each inclusion compound was calculated from the atomic coordinates with the program Free Volume10 in the Cerius2 software package (version 4.0).11 Calculation involved the rolling of a spherical probe along the interior surface; the probe radius was 0.7 Å.6 The following values were used for the atomic radii: H⊕=⊕1.20 Å, C⊕=⊕1.70 Å, O⊕=⊕1.60 Å, N⊕=⊕1.65 Å and Cl⊕=⊕1.75 Å.Acknowledgements
This work was supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture, Japan.References
- (a) K. Miki, A. Masui, N. Kasai, M. Miyata, M. Shibakami and K. Takemoto, J. Am. Chem. Soc., 1988, 110, 6594 CrossRef CAS; (b) K. Miki, N. Kasai, M. Shibakami, K. Takemoto and M. Miyata, J. Chem. Soc., Chem. Commun., 1991, 1757 RSC; (c) M. Miyata, K. Sada, K. Hirayama, Y. Yasuda and K. Miki, Supramol. Chem., 1993, 2, 283 CrossRef CAS; (d) K. Nakano, K. Sada and M. Miyata, Chem. Commun., 1996, 989 RSC; (e) K. Nakano, K. Sada and M. Miyata, Prog. Colloid Polym. Sci., 1997, 106, 249 CrossRef CAS; (f) K. Nakano, M. Katsuta, K. Sada and M. Miyata, CrystEngComm, 2001, 11; http://www.rsc.org/ej/ce/2001/b009361k/index.htm Search PubMed.
- P. L. Johnson and J. P. Schaefer, Acta Crystallogr., Sect. B, 1972, 28, 3083 CrossRef CAS; E. L. Jones and L. R. Nassimbeni, Acta Crystallogr., Sect. B, 1990, 46, 399 CrossRef.
- M. Shibakami and A. Sekiya, J. Inclusion Phenom., 1994, 18, 397 CrossRef CAS; M. Shibakami, M. Tamura and A. Sekiya, J. Inclusion Phenom., 1995, 22, 155 CrossRef; M. Shibakami, M. Tamura and A. Sekiya, J. Inclusion Phenom., 1995, 22, 299 CrossRef; M. Shibakami, M. Tamura and A. Sekiya, J. Am. Chem. Soc., 1995, 117, 4499 CrossRef.
- (a) M. R. Caira, L. R. Nassimbeni and J. L. Scott, J. Chem. Soc., Chem. Commun., 1993, 612 RSC; (b) M. R. Caira, L. R. Nassimbeni and J. L. Scott, J. Chem. Soc., Perkin Trans. 2, 1994, 623 RSC; (c) M. R. Caira, L. R. Nassimbeni and J. L. Scott, J. Chem. Soc., Perkin Trans. 2, 1995, 495 Search PubMed; (d) J. L. Scott, J. Chem. Soc., Perkin Trans. 2, 1995, 495 RSC; (e) P. Dastidar, CrystEngComm, 2000, 8 Search PubMed.
- M. Miyata and K. Sada, in Comprehensive Supramolecular Chemistry, Solid-state Supramolecular Chemistry: Crystal Engineering, ed. D. D. MacNichol, F. Toda and R. Bishop, Pergamon, Oxford, 1996, vol. 6, pp. 147–176 Search PubMed.
- K. Nakano, K. Sada, Y. Kurozumi and M. Miyata, Chem. Eur. J., 2001, 7, 209 CrossRef CAS.
- M. Shibakami and A. Sekiya, J. Chem. Soc., Chem. Commun., 1994, 429 RSC.
- SIR92, A Program for Automatic Solution of Crystal Structures by Direct Methods: A. Altomare, M. Cascarano, C. Giacovazzo, A. Guagliardi, M. C. Burla, G. Polidori and M. Camalli, J. Appl. Crystallogr., 1994, 27, 435 Search PubMed.
- TeXsan, X-Ray Structure Analysis Package , Molecular Structure Corporation, The Woodlands, TX, 1985 Search PubMed.
- R. Voorintholt, M. T. Kosters, G. Vegter, G. Vriend and W. G. J. Hol, J. Mol. Graphics, 1989, 7, 243 CrossRef CAS PubMed.
- Cerius2, Molecular Simulation Software , Molecular Simulations Inc., San Diego, CA, 1998 Search PubMed.
| This journal is © The Royal Society of Chemistry 2001 |