Docking-guided identification of protein hosts for GFP chromophore-like ligands†
Abstract
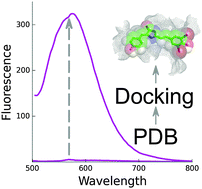
Synthetic analogs of the Green Fluorescent Protein (GFP) chromophore emerge as promising fluorogenic dyes for labeling in living systems. Here, we report the computational identification of protein hosts capable of binding to and enhancing fluorescence of GFP chromophore derivatives. Automated docking of GFP-like chromophores to over 3000 crystal structures of Escherichia coli proteins available in the Protein Data Bank allowed the identification of a set of candidate proteins. Four of these proteins were tested experimentally in vitro for binding with the GFP chromophore and its red-shifted Kaede chromophore-like analogs. Two proteins were found to possess sub-micromolar affinity for some Kaede-like chromophores and activate fluorescence of these fluorogens.
- This article is part of the themed collection: Shape-Responsive Fluorophores


 Please wait while we load your content...
Please wait while we load your content...