Comparison of three competing dynamic force spectroscopy models to study binding forces of amyloid-β (1–42)†
Abstract
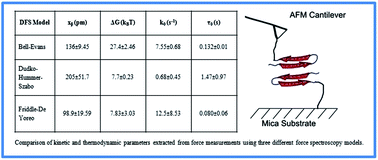
We performed single molecule dynamic force spectroscopy experiments to study the dimerization of two amyloid-β (1–42) peptides and compared three different theoretical models used to fit experimental data: Bell–Evans, Dudko–Hummer–Szabo, and Friddle–De Yoreo. Using these models we extracted values of the dissociation rate at zero force, k0, and height and the width of the energy barrier, ΔG and xβ. We show the importance of including the effect of the linker molecule. All three models corrected for the linker effect give comparable results for xβ and show more discrepancy for k0 and ΔG values, ΔG parameter correlates well between Dudko–Hummer–Szabo and Friddle–De Yoreo models but differs for the Bell–Evans model.

 Please wait while we load your content...
Please wait while we load your content...