Structural basis for recognition of bacterial cell wall teichoic acid by pseudo-symmetric SH3b-like repeats of a viral peptidoglycan hydrolase†
Abstract
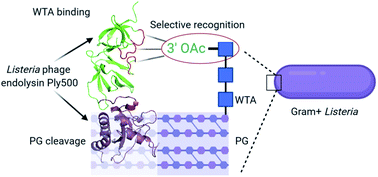
Endolysins are bacteriophage-encoded peptidoglycan hydrolases targeting the cell wall of host bacteria via their cell wall-binding domains (CBDs). The molecular basis for selective recognition of surface carbohydrate ligands by CBDs remains elusive. Here, we describe, in atomic detail, the interaction between the Listeria phage endolysin domain CBD500 and its cell wall teichoic acid (WTA) ligands. We show that 3′O-acetylated GlcNAc residues integrated into the WTA polymer chain are the key epitope recognized by a CBD binding cavity located at the interface of tandem copies of beta-barrel, pseudo-symmetric SH3b-like repeats. This cavity consists of multiple aromatic residues making extensive interactions with two GlcNAc acetyl groups via hydrogen bonds and van der Waals contacts, while permitting the docking of the diastereomorphic ligands. Our multidisciplinary approach tackled an extremely challenging protein–glycopolymer complex and delineated a previously unknown recognition mechanism by which a phage endolysin specifically recognizes and targets WTA, suggesting an adaptable model for regulation of endolysin specificity.
- This article is part of the themed collection: 2020 Chemical Science HOT Article Collection


 Please wait while we load your content...
Please wait while we load your content...