A bright FIT-PNA hybridization probe for the hybridization state specific analysis of a C → U RNA edit via FRET in a binary system†
Abstract
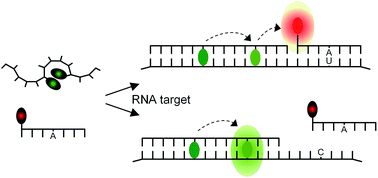
Oligonucleotide probes that show enhanced fluorescence upon nucleic acid hybridization enable the detection and visualization of specific mRNA molecules, in vitro and in cellulo. A challenging problem is the analysis of single nucleotide alterations that occur, for example, when cellular mRNA is subject to C → U editing. Given the length required for uniqueness of the targeted segment, the commonly used probes do not provide the level of sequence specificity needed to discriminate single base mismatched hybridization. Herein we introduce a binary probe system based on fluorescence resonance energy transfer (FRET) that distinguishes three possible states i.e. (i) absence of target, (ii) presence of edited (matched) and (iii) unedited (single base mismatched) target. To address the shortcomings of read-out via FRET, we designed donor probes that avoid bleed through into the acceptor channel and nevertheless provide a high intensity of FRET signaling. We show the combined use of thiazole orange (TO) and an oxazolopyridine analogue (JO), linked as base surrogates in modified PNA FIT-probes that serve as FRET donor for a second, near-infrared (NIR)-labeled strand. In absence of target, donor emission is low and FRET cannot occur in lieu of the lacking co-alignment of probes. Hybridization of the TO/JO-PNA FIT-probe with the (unedited RNA) target leads to high brightness of emission at 540 nm. Co-alignment of the NIR-acceptor strand ensues from recognition of edited RNA inducing emission at 690 nm. We show imaging of mRNA in fixed and live cells and discuss the homogeneous detection and intracellular imaging of a single nucleotide mRNA edit used by nature to post-transcriptionally modify the function of the Glycine Receptor (GlyR).


 Please wait while we load your content...
Please wait while we load your content...