The relative stability of trpzip1 and its mutants determined by computation and experiment†
Abstract
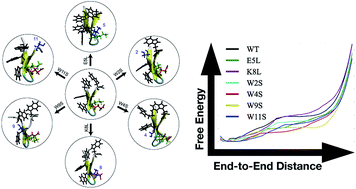
Six mutants of the tryptophan zipper peptide trpzip1 have been computationally and experimentally characterized. We determine the varying roles in secondary structure stability of specific residues through a mutation assay. Four of the mutations directly effect the Trp–Trp interactions and two of the mutations target the salt bridge between Glu5 and Lys8. CD spectra and thermal unfolding are used to determine the secondary structure and stability of the mutants compared to the wildtype peptide. Adaptive steered molecular dynamics has been used to obtain the energetics of the unfolding pathways of the mutations. The hydrogen bonding patterns and side-chain interactions over the course of unfolding have also been calculated and compared to wildtype trpzip1. The key finding from this work is the importance of a stabilizing non-native salt bridge pair present in the K8L mutation.


 Please wait while we load your content...
Please wait while we load your content...